Translate this page into:
MicroRNA Expression and Target Prediction in Children with Nephrotic Syndrome
Corresponding author: Mohanapriya Chinambedu Dandapani, Department of Human Genetics, SRIHER, Chennai - 600116, Tamil Nadu, India. E-mail: mohanapriya@sriramachandra.edu.in
-
Received: ,
Accepted: ,
How to cite this article: Charmine P, Venkatesan V, Geminiganesan S, Nammalwar BR, Dandapani MC. MicroRNA Expression and Target Prediction in Children with Nephrotic Syndrome. Indian J Nephrol. 2025;35:59-63. doi: 10.25259/ijn_47_23
Abstract
Background:
Nephrotic syndrome is a common cause of kidney diseases in children. Many studies have examined the association of microRNAs playing potential roles in many pathophysiological functions. We investigated the expression pattern of the microRNAs miR-17-5P, miR-155p, miR-424-5p in children with steroid sensitive nephrotic syndrome (SSNS) and steroid resistance nephrotic syndrome (SRNS), along with the healthy subjects.
Materials and Methods:
Total RNA was isolated from the urine samples from the three groups (SSNS n = 100, SRNS n = 100, and healthy control group n = 100). Bioinformatics tools such as miRWalk and miR-Tar link were used in predicting targets for the microRNAs. Online database and g profiler software are used to evaluate the targets based on the biological functions. The expression pattern for the candidate microRNAs was carried out using quantitative real time polymerase chain reaction (RT-PCR) equipment.
Results:
miR-424 and miR-155 were upregulated in SRNS group while miR-17 was downregulated in SRNS group. miR-424-5p and miR-155p was up regulated in SRNS group while miR-17-5p was downregulated.
Conclusion:
Combined analysis of gene expression along with studied candidate microRNAs can give better understanding of the pathogenesis of childhood nephrotic syndrome.
Keywords
Expression analysis
g profile
microRNAs
miR Tar Link
miRWalk
Nephrotic syndrome
Introduction
Nephrotic syndrome is a common kidney syndrome in children.1-4 Certain microRNAs have been experimentally validated in order to identify and select the specific function related to this condition.5 Almost 80% of children with idiopathic nephrotic syndrome show remission of proteinuria with corticosteroids (steroid sensitive), but nearly 40%–60% may tend to experience intermittent relapses.6-8 Steroid resistance denotes the absence of remission following at least 4 weeks of prednisone therapy.9 While about 30% patients with steroid resistance have an underlying genetic etiology, the exact disease mechanisms for others remain elusive. Epigenetic modification of gene expression, including expression of noncoding RNA (microRNAs), might provide an etiological clue in patients without a definitive genetic etiology. Further research is needed to fully comprehend the implications of this discovery and explore potential interventions or treatments.10 Our study focuses on idiopathic nephrotic syndrome in children with steroid sensitive nephrotic syndrome (SSNS) and steroid resistant nephrotic syndrome (SRNS).
MicroRNAs (miRNAs), which regulate target genes in the post-transcriptional processes and play crucial roles in many disorders, are about 20 nucleotides in length. miRNAs have been demonstrated to function as noninvasive biomarkers in a variety of diseases, including renal disorders, in serum and urine.11,12
Both microRNAs (miRNAs) and RNA binding proteins (RBPs) are crucial transcriptional regulators of gene expression. These two factors also act to reduce translation by translation repression or miRNA cleavage.13 They are found to be a biomarker not only in diseases such as viral infections, cardiovascular disorders, diabetes, and cancer, but also in kidney diseases.14
Kidney disease has been discovered to be significantly influenced by miR-155. It has been linked to a number of pathological processes, including fibrosis, immune response regulation, and inflammation, all of which are crucial for the onset and progression of kidney disorders.6 A study by Wang et al., has compared expression pattern for miR-424 between the nondiabetics along with the diabetic nephropathy patients and it was observed that the expression of miR-424 in the kidneys of diabetic nephropathy patients was considerably higher. Additionally, inhibiting miR-424 decreased kidney damage and fibrosis in vitro, indicating that miR-424 may be involved in the onset and progression of diabetic nephropathy in people.7 When compared to healthy controls, the researchers discovered miR-17 was noticeably elevated in the glomeruli of patients with membranous nephropathy. miR-17 overexpression increased albuminuria and podocyte death in cultured podocytes, pointing to a potential function for miR-17 in the onset and progression of membranous nephropathy.8
The present study investigated the differential expression of miR-17-5p, miR-155p, miR- 424 -5p, with miR-484-5p as endogenous control in urine samples between the study groups.
Materials and Methods
The study was conducted at Sri Ramachandra Institute of Higher Education and Research and the samples were collected from the Department of Nephrology, SRIHER, Chennai, India. The children who have met the criteria were enrolled for the study are within the age group 1–12 years. The sampling for the SRNS was taken before the starting of calcineurin inhibitors and then for the SSNS group, it was taken during the relapse. The study included 300 participants, 200 cases with SSNS and SRNS, and 100 age-matched controls. Informed consent was obtained from the parents or guardians of all participants. Ethics committee approval was obtained from the Institutional Ethics committee at Sri Ramachandra Institute of Higher Education and Research [IEC-NI/21/FEB/77/2].
Study design
Patients with SSNS were enrolled during relapse prior to initiating therapy. The SRNS subjects (defined as non response to oral prednisolone at 4 weeks of therapy) were enrolled before initiating therapy with calcineurin inhibitors.
Secondary NS and familial NS were excluded.
Computational prediction for microRNAs using online database
MicroRNA prediction tools such as miRWalk and miR Target link human were used to select the specific targets for the study. The selected microRNAs were experimentally validated and analysed using the g profiler software in order to identify and select the specific function related to this disease condition.5
Sample collection and RNA purification
6mL of urine was collected in a sterile container. The samples are pelleted by centrifugation for 20 min at 3,000 rpm. The urinary pellet was lysed with RNA lysis buffer and it was stored in the deep freezer at 80°C for further purpose. MicroRNA isolation was carried out for all the samples from the urinary pellet by using the MiRNeasy Mini Kit, and the procedure was performed according to the manufacturer’s protocol. Urine samples were mixed thoroughly with an equal volume of miRNeasy Serum/Plasma Spike-In Control, incubated for 2–3 min at room temperature, and then analyzed and centrifuged at 12,000g for 15 min at 4°C. After transferring the upper aqueous phase to a new collecting tube, 1.5 L of 100% ethanol were added. A 2mL collection tube containing 700 L of material was pipetted into a RNeasy Min Elute spin column and spun at 8000g for 15 s at room temperature. The supernatant was discarded. The RNeasy Min Elute spin column received about 500 L of elution buffer, and it was centrifuged at the same speed. The RNeasy Min Elute spin column was filled with approximately 500 µL of 80% ethanol and centrifuged to elute the RNA.
Quantification of purified RNA
The concentrations and purity of the isolated RNA samples were quantified using NanoDrop 1000 equipment for all the samples by measuring the absorbance at 260 and 280 nm. The ratio of 1.7–2.0 and around 50 ng concentration was observed in all the purified samples denoting the good quality of RNA. All the 300 samples were checked for quality and quantity and they were found to be good and with adequate proportion for expression study.
cDNA conversion and microRNA expression using quantitative real time PCR
The cDNA conversion was performed using the Taqman advanced cDNA synthesis kit and the cycling conditions for poly A tailing is polyadenylation 37°C for 45 min, ligation 16°C for 60 min, reverse transcription 42°C for 15 min; miR-amp reaction was enzyme activation 95°C for 5 min, deanture 95°C for 3 s, annealing/extension 60°C 30 s, stop reaction 99°C for 10 min and hold at 4°C. The converted cDNA templates were further diluted to proceed for microRNA expression analysis using Rotar Gene Q equipment (Qiagen) for the selected miRNAs miR-17-5P, miR-155p, and miR-424-5P. The RT-PCR reaction components include the Taqman advanced master mix, targeted probe, and sterile water. The RT-PCR cycling condition mentioned was elaborated. Statistical (t-test) and bioinformatics analysis were performed.
Results
Target prediction analysis
The miRtarlink has revealed that miR-155p, miR-424-5p, and miR-17-5P are found to have stronger predication connected with the diseases pathogenesis and are represented in Figure 1 and miRWalk is represented in Table 1. The targeted microRNAs were also analyzed for the various pathways that are associated with the nephrotic syndrome. A detailed analysis involving various functions of candidate miRNAs has been broadly represented with their unique IDs in Table 1.
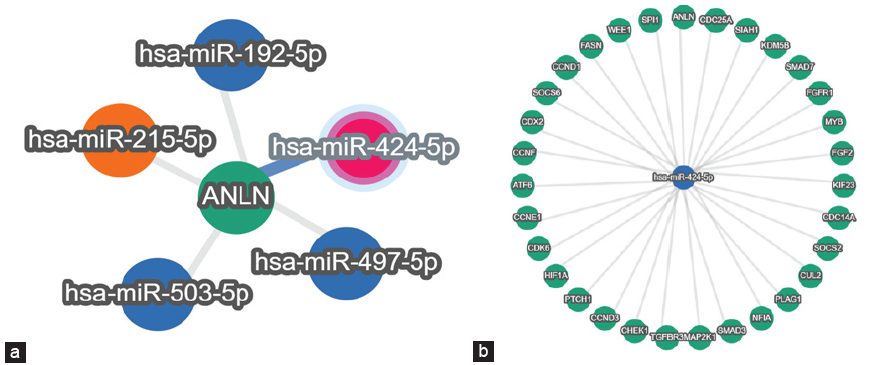
- (a) Represents the gene ANLN and its targeting microRNAs. (b) Represents the microRNA (miR-424-5p) and its targeting genes.
| microRNAs | Predicted Targets |
|---|---|
| has-miR-424-5p | VEGFA |
| has-miR-17-5p | IL8 |
| has-miR-155-5p | IL8, AGTR1, TRPS1, NR3C1 |
| Description | Term Id | Corrected P Value |
|---|---|---|
| Molecular Function | ||
| Cytokine receptor binding | GO: 0005126 | 3.952×10-4 |
| Signaling receptor binding | GO: 0005102 | 1.025×10-3 |
| Vascular endothelial growth factor receptor binding | GO: 0005172 | 1.902×10-3 |
| Biological Process | ||
| Cell migration | GO: 0016477 | 1.620×10-2 |
| Localization of cell | GO: 0051674 | 2.953×10-2 |
| Cell motility | GO: 0048870 | 2.953×10-2 |
| Cellular Components | ||
| Platelet alpha granule lumen | GO: 0031093 | 3.905×10-2 |
| Reactome Pathway | ||
| Nephrin family interactions | REAC: R-HSA-37375 | 2.911×10-2 |
| Wiki Pathway | ||
| Nephrotic syndrome | WP: WP4758 | 9.984×10-4 |
| HP | ||
| Abnormal nephron morphology | HP: 001257 | 1.165×10-3 |
| Focal segmental glomerulosclerosis | HP: 0000097 | 1.715×10-2 |
| Abnormal renal glomerulus morphology | HP: 0000095 | 2.653×10-3 |
| Abnormal renal glomerulus morphology | HP: 0000095 | 3.929×10-3 |
| Proteinuria | HP: 0000093 | 5.157×10-3 |
| Glomerular sclerosis | HP: 0000096 | 5.157×10-3 |
| Abnormal urine protein level | HP: 0020129 | 5.649×10-3 |
RNA: Ribonucleic acid, VEGFA: Vascular endothelial growth factor A, AGTR: Angiotensin II Receptor Type 1, TRPS: Trichorhinophalangeal Syndrome, NR3C: Nuclear Receptor Subfamily 3 Group C Member (also known as Glucocorticoid Receptor), HP: Haptoglobin, REAC: Ras-Responsive Element-Binding Protein (also known as ATF2 - Activating Transcription Factor 2), R-HSA: Reactome Database (R) Stable identifier corresponding species (Homosapiens (HSA), WP: Wingless-related Pathway).
MicroRNA expression analysis
The basic demographic characteristics are shown in Table 1. In comparison to the control group, it was observed that miR-17, miR-424, and miR-155 were elevated in SSNS and SRNS as shown in Figure 2. Further, the microRNAs were compared within the case groups (SSNS, SRNS) to identify the fold change and found that miR-424 and miR-155 are upregualated (two-fold increase) in SRNS group while miR-17 is found to be downregulated in SRNS group as shown in Figure 3 and Table 2.
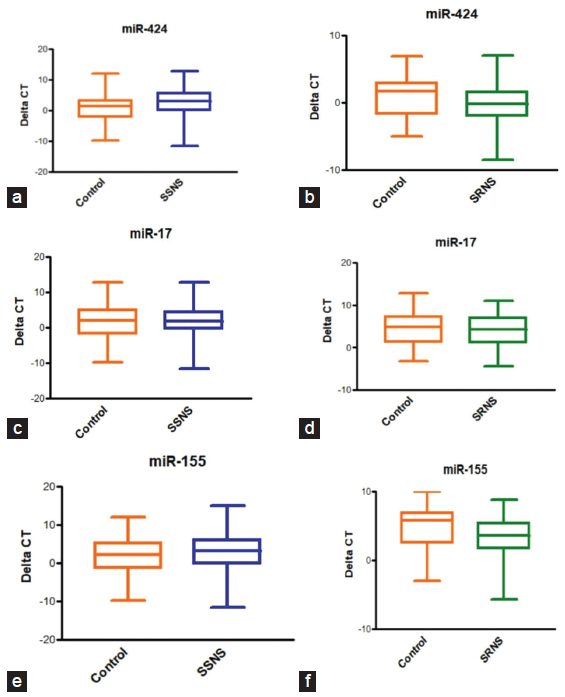
- (a-f) Urinary microRNA expression of miR-155, miR-424 and miR-17 in children with SSNS and SRNS. SSNS: Steriod sensitive nephrotic syndrome, SRNS: Steriod resistant nephrotic syndrome.
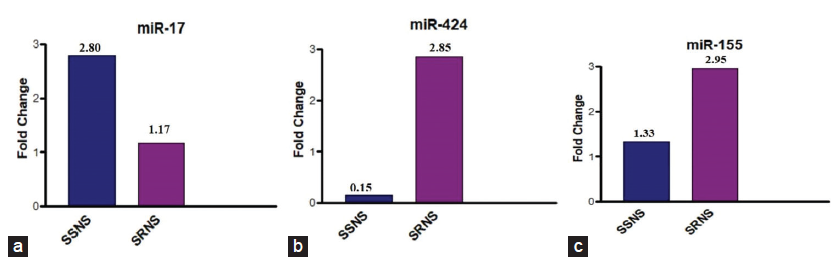
- (a-c) Bar diagrams showing fold change in the expression levels of miR-424.,miR-17 and miR-155 among the cases. SSNS: Sensitive nephrotic syndrome, SRNS: Steroid resistant nephrotic syndrome.
| miRNA | Fold change | P value |
|---|---|---|
| miR-424 |
SSNS = 0.12 SRNS = 2.80 |
SSNS = <.0001 SRNS = 0.00056 |
| miR-17 |
SSNS = 2.80 SRNS = 1.72 |
SSNS = 0.001779 SRNS = 0.329064 |
| miR-155 |
SSNS = 1.33 SRNS = 2.95 |
SSNS = 0.179375 SRNS = 0.0009 |
RNA: Ribonucleic acid, SSNS: Steroid sensitive nephrotic syndrome, SRNS: Steroid resistant nephrotic syndrome.
Discussion
Many studies have reported the importance of microRNAs and its potential roles in apoptosis, cell differentiation and cell proliferation with the inclusion of the kidney disease. Differential expression miRNAs studies has been helpful in genome-wide profiling studies.10-14 In kidney injury, miR-21 and miR-155 were found to show decreased level of expression in blood and urine samples.15 By using a micro-RNA array, Luo et al. showed that miRNAs mir30a05p, mir-151-3p, and miR-191 showed high level of expression in urine and serum samples in NS children when compared to the healthy individuals.16
In this study, we investigated the expression of miR-17 on the pathogenesis of nephrotic syndrome. The expression pattern observed for miR-17 was downregulated in our study, while a study by Zang et al. (2020), showed that mir-17-5p was found to be significantly increased in childhood nephrotic syndrome when compared with the healthy subjects.17 The miR-17 deregulation has been seen in the kidneys of nephrotic syndrome patients in numerous investigations. The authors hypothesized that elevated miR-17 expression may aid in FSGS development by encouraging podocyte damage and dysfunction.17
Our findings revealed that miR-155 expression was significantly upregulated in the glomeruli of patients with active minimal change disease (MCD), a common cause of nephrotic syndrome in children. These results are consistent with previous studies by Ramezani, et al.,18 and Zhang et al. (2020).19
Our findings are in contrast to the study by Shaffi SK, et al. that has investigated the expression levels of miR-424 in urine and plasma samples renal parenchymal diseases which has shown down regulation.20
The limitation of the study is that the biopsy correlation with miRNA expression was not performed because our study was not focused on the kidney biopsy results; no correlation analysis was performed for the data on the kidney biopsy.
This was the first study to report the expression pattern of the microRNAs which specifically plays an important role in the podocyte junction of actin cytoskeleton. miR-424-5p and miR-155p is upregulated in SRNS group while miR-17-5p is downregulated. Combined analysis of gene expression along with the studied candidate microRNAs can give a better understanding in bringing out the knowledge in the childhood nephrotic syndrome in future studies.
Financial support and sponsorship
The study was supported by the Department of Biotechnology, Ministry of Science and Technology (Ref No: BT/PR30523/BIC/101/1121/2018).
Conflicts of interest
There are no conflicts of interest.
References
- Management of nephrotic syndrome in the pregnant patient. J Reprod Med. 2016;61:557-61.
- [PubMed] [Google Scholar]
- An update on LDL apheresis for nephrotic syndrome. Pediatr Nephrol. 2019;34:1655-69.
- [CrossRef] [PubMed] [Google Scholar]
- Idiopathic nephrotic syndrome: Characteristics and identification of prognostic factors. J Clin Med. 2018;7:265.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- RWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Carolina Tapia; Khalid Bashir Nephrotic Syndrome May 29, 2023.
- Upregulation of microrna-424 relieved diabetic nephropathy by targeting Rictor through mtor Complex2/Protein Kinase B signaling. J Cell Physiol. 2019;234(7):11646-11653.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNAs as biomarkers for nephrotic syndrome. Int J Mol Sci. 2020;23(22):88.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66.
- [CrossRef] [PubMed] [Google Scholar]
- Protecting podocytes: A key target for therapy of focal segmental glomerulosclerosis. Am J Nephrol. 2018;47(Suppl 1):14-29.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Changing etiologies of unexplained adult nephrotic syndrome: A comparisonof renal biopsy findings from 1976–1979 and 1995–1997. Am J Kidney Dis. 1997;30:621-31.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNAs as biomarkers for nephrotic syndrome. Int J Mol Sci. 2021;22:88.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci. 2012;129:256-67.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Increased serum and urinarymicroRNAs in children with idiopathic nephrotic syndrome. Clin Chem. 2013;59:658-66.
- [CrossRef] [PubMed] [Google Scholar]
- Role of microRNA-17-5p in the pathogenesis of pediatric nephrotic syndrome and related mechanisms. Zhongguo Dang Dai Er Ke Za Zhi. 2020;22:958-963.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: a pilot study. Eur J Clin Invest. 2015;45:394-404.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miR-155-5p Implicates in the pathogenesis of renal fibrosis via targeting SOCS1 and SOCS6. Oxid Med Cell Longev 2020:6263921.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Role of MicroRNAs in renal parenchymal diseases-a new dimension. Int J Mol Sci. 2018;19:1797.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]








