Translate this page into:
Distribution of Virulence Factors According to Antibiotic Susceptibility among Escherichia coli Isolated from Urinary Tract Infection
Address for correspondence: Dr. S. Derakhshan, Department of Microbiology, Faculty of Medicine, Kurdistan University of Medical Sciences, Sanandaj, Iran. E-mail: s.derakhshan@muk.ac.ir
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Medknow Publications & Media Pvt Ltd and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Escherichia coli is the major causative pathogen of urinary tract infection (UTI) in humans. Virulence and drug resistance play important roles in the pathogenesis of E. coli infections. The aims were to investigate the presence of uropathogenic virulence genes and to evaluate a relationship between antibiotic resistance and virulence in E. coli from UTI. A total of 132 E. coli were collected between April and June 2015 in two hospitals of Sanandaj, Iran. Isolates were examined for susceptibility to 16 antibiotic disks using the disk diffusion method and for possession of virulence genes by polymerase chain reaction. Associations between antimicrobial resistance and virulence genes were investigated. A P < 0.05 was considered significant. Of the 132 isolates, the most prevalent virulence gene was pap (31.1%), followed by cnf (28.8%), hly (16.7%), and afa (10.6%). Different patterns of virulence genes were identified. A significant association was detected between the simultaneous presence of hly and pap. The most effective antibiotics were nitrofurantoin, cefoxitin, and imipenem and the least effective were ampicillin, trimethoprim-sulfamethoxazole, and cefotaxime. An association was seen between the presence of cnf and susceptibility to the certain antibiotics, whereas strains with a reduced susceptibility to the certain antibiotics were associated with a significantly increased prevalence of afa and hly (P < 0.05). These findings suggest a correlation between the presence of virulence gene and resistance in E. coli strains from UTI. The results indicate that there is a need for surveillance programs to monitor drug resistance in pathogenic E. coli.
Keywords
Antimicrobial agents
Escherichia coli
resistance
urinary tract infection
virulence
Introduction
Pathogenic strains of Escherichia Coli have the potential to cause a wide variety of infectious diseases, including neonatal meningitis, septicemia, intestinal tract infection, and urinary tract infections (UTIs).[1] UTI is one of the most frequent bacterial infections, affecting both inpatients and outpatients, and E. coli is the major causative pathogen.[2] Due to anatomical differences and the hormonal milieu of the urinary tract, the probability of developing a UTI in women is significantly more than men. It is estimated that around 50%–60% of women will experience a UTI during their lifetime.[3]
UTI caused by E. coli strains remains an important health problem in many countries and leads to considerable morbidity costs.[145] These strains encode various virulence factors such as toxins, capsules, invasins, and adhesins, which contribute to enhanced pathogenicity. The severity of UTI reflects the virulence profile of the infecting strain, and the frequency of expression of virulence factors is higher in more pathogenic strains.[6]
Pyelonephritis-associated pilus (Pap), afimbrial adehsin (Afa), α-hemolysin (HlyA), and cytotoxic necrotizing factor 1 (CNF1) are among the most important virulence factors of E. coli involved in the development of UTI.[789] Pap is one of the most commonly found adhesins. Binding of P-fimbrial adhesin to the cell receptors of renal tissue leads to mucosal inflammation and tissue damage.[7] Afa has been implicated in the occurrence of recurrent and chronic UTIs.[10] Apart from adhesins, exotoxins such as HlyA and CNF1 are also implicated in the pathogenesis of UTIs. HlyA encoded by hly is the most important secreted virulence factor of E. coli isolated from UTI. This toxin is able to lyse host cells for crossing of the mucosal barriers, having access to host nutrients and iron stores, and damaging effector immune cells such as T-lymphocytes and neutrophils. CNF1 has been shown to have a role in dissemination of strains in the urinary tract. This toxin causes bladder cell exfoliation and increases bacterial access to the underlying tissue.[8]
Another problem is antibiotic resistance of the E. coli strains. The emergence of resistance limits the utility of older agents, such as trimethoprim-sulfamethoxazole (SXT), and increases reliance on newer broad-spectrum agents, such as extended-spectrum cephalosporins and fluoroquinolones. Unfortunately, emerging resistance now threatens the use of these newer agents as well.[111213] Resistance to antimicrobial agents is often associated with the spread of transmissible plasmids, which may also carry virulence determinants.[14] Some studies showed that among clinical isolates of E. coli, the production of virulence factors is negatively associated with resistance to some antibiotics.[1516] On the other hand, such phenomenon was not observed in other studies.[171819] The acquisition of resistance and virulent traits may provide a benefit for the survival of microorganism. This situation may lead to ecological changes and domination of virulent antibiotic-resistant bacteria in the environment.[14]
Hence, the aims of this study were to determine the presence of the most important virulence genes involved in the development of UTI including pap, afa, cnf-1, and hly[789] among E. coli isolated from UTI and to evaluate a possible association between virulence factors and susceptibility to antimicrobial agents.
Materials and Methods
Bacterial isolates and identification
In this cross-sectional study, from April to June 2015, 132 consecutive nonduplicate E. coli were isolated from patients with UTI admitted to Besat and Tohid Tertiary Hospitals in Sanandaj, Iran. Sanandaj is the center of Kurdistan Province in the west of Iran, with a population of more than 300,000. The Besat and Tohid Hospitals are referral and teaching hospitals, which are affiliated to Kurdistan University of Medical Sciences. UTI was defined according to the 2015 European Association of Urology guidelines.[20] E. coli isolates were identified according to the standard bacteriological and biochemical tests[21] including Gram staining, fermentation of lactose, motility, ability to produce indole, and lysine decarboxylation. All bacterial isolates were preserved at −70°C in Trypticase soy broth (Quelab, New Mexico, USA), containing 15% v/v glycerol for further investigations.
Antimicrobial susceptibility testing
Susceptibility of E. coli isolates was determined to 16 antibiotics using the disk diffusion method on Mueller-Hinton agar plates (Merck, Germany) as recommended by the 2014 Clinical and Laboratory Standards Institute guidelines.[22] The following antibiotic disks (Rosco company, Denmark) were tested: ampicillin (AM) (10 μg), cefotaxime (CTX) (30 μg), ceftazidime (CAZ) (30 μg), imipenem (IPM) (10 μg), amoxicillin/clavulanic acid (AMC) (20/10 μg), aztreonam (AZT) (30 μg), ciprofloxacin (CP) (5 μg), tetracycline (TE) (30 μg), SXT, gentamicin (GM) (10 μg), cefepime (FEP) (30 μg), cefoxitin (CFO) (30 μg), amikacin (AN) (30 μg), norfloxacin (NOR) (10 μg), nalidixic acid (NA) (30 μg), and nitrofurantoin (FM) (300 μg). E. coli ATCC 25922 was used as a quality control strain.
DNA extraction and detection of virulence factors by polymerase chain reaction
Genomic DNA was extracted from E. coli strains by the freeze-thaw method and used as the template for polymerase chain reaction (PCR) reactions.[23] For DNA extraction, bacterial pellets prepared from 1.5 ml of an overnight culture in brain–heart infusion broth (Quelab, New Mexico, USA) were suspended in 200 μl sterile distilled water, and the suspensions were heated at 100°C for 10 min. The suspensions were then immediately placed on ice for 5 min. Samples taken through a total of three cycles of freezing-thawing were centrifuged, and the supernatants were stored at −20°C as DNA template stocks.
Detection of virulence genes was carried out by PCR. Amplification of pap, afa, cnf-1, and hly was performed using published primer pairs (SinaClon, Iran) as follows: 5′-AACAAGGATAAGCACTGTTCTGGCT-3′, 5′-ACCA TATAAGCGGTCATTCCCGTCA-3′ (for hly, amplicon size: 1177 bp);[24] 5′-AAGATGGAGTTTCCTATGCAGGAG-3′, 5′-CATTCAGAGTCCTGCCCTCATTATT-3′ (for cnf, amplicon size: 498 bp);[24] 5′-GCTG GGCAGCAAACTGATAACTCTC-3′, 5′-CATCAAGCTGTTTGTTCGTCCGCCG-3′ (for afa, amplicon size: 750 bp);[25] and 5′-GA CGGCTGTACTGCAGGGTGTGGCG-3′, 5′-ATATCCTTTCTGCAGGGATGCAATA-3′ (for pap, amplicon size: 328 bp).[25]
Amplification reactions were done in a total volume of 25 μl containing 3 μl DNA extract, 1 U of Taq DNA polymerase, 0.4 μM of each primer, 1X reaction buffer, 1.5 mM MgCl2, and 200 μM of each dNTP (SinaClon, Iran). The PCR was performed in a thermal cycler (Eppendorf, Germany) under the following conditions: initial denaturation of 5 min at 94°C followed by 35 cycles of denaturation of 1 min at 94°C, annealing of 1 min at 65°C, extension of 1 min at 72°C, and a final extension of 7 min at 72°C. Conditions were the same for all genes.
The amplified products were separated on a 1% agarose gel (SinaClon) in 0.5X tris-borate EDTA buffer alongside an appropriate molecular size marker (100 bp Plus DNA ladder, SinaClon). The amplified products were visualized after staining with DNA safe stain (SinaClon) and photographed using a UV transillumination imaging system. Positive controls were kindly given by Dr. S. Najar Peerayeh (Tarbiat Modares University, Iran) and Dr. A. Rashki (University of Zabol, Iran).
Statistical analyses
Data were analyzed using SPSS software version 16.0 (SPSS, Chicago, IL, USA). The relationship between virulence factors and antibiotic susceptibility was determined using Pearson's Chi-square test or Fisher's exact test. To facilitate our analysis, the isolates showing intermediate susceptibility were grouped with the resistant strains. A P< 0.05 was considered significant.
Results
A total of 132 E. coli strains were collected from patients with UTI between April and June 2015. The average age of the patients was 35.7 years; the oldest patient was 93 years (one patient) and four 1-year-old children were the youngest patients. Among the 132 isolates, 107 isolates (81.1%) were from females and 25 (18.9%) were from males. Forty-seven (35.6%) of the 132 isolates were from hospitalized patients and 85 (64.4%) were from outpatients.
Antimicrobial susceptibility
Antimicrobial susceptibility results showed that all isolates (97%) were susceptible to FM, except for four isolates. Of the 132 isolates, 122 (92.4%) were susceptible to CFO, 116 (87.9%) to IPM, 99 (75%) to AN, and 95 (72%) to AMC. The susceptibility rate of isolates to 16 antimicrobial agents is presented in Table 1. Compared with the isolates from inpatients, except for AMC, FM, IPM, AN, and GM, the frequency of susceptibility to the antimicrobial agents was higher or similar in the outpatients isolates [Table 1].

Distribution of virulence genes
Prevalence of virulence genes was analyzed by PCR. Of the 132 isolates, the pap gene was found in 41 (31.1%) isolates, the cnf in 38 (28.8%), the hly in 22 (16.7%), and the afa in 14 (10.6%) isolates. Fifty-seven strains were negative for all the studied virulence genes. The prevalence of virulence genes was higher in males, except for afa, which was detected in a higher prevalence in females than in males (11.2% vs. 8%) [Table 2]. Among the studied virulence genes, only pap showed a meaningful difference of distribution according to sex group (P = 0.01).

Except pap gene, that its prevalence in the inpatient group was higher than outpatient group (16/47 [34%] vs. 25/85 [29.4%]), the prevalence of other virulence genes was higher in the outpatient group. The hly, cnf, and afa were detected in 7 (14.9%), 12 (25.5%), and 2 (4.3%) of the 47 strains collected from inpatients and in 15 (17.6%), 26 (30.6%), and 12 (14.1%) of the 85 strains collected from outpatients, respectively. However, the prevalence of virulence genes was not significantly different between the two groups (P > 0.05).
To identify phenotypes of the isolates, the prevalence of profiles of the virulence genes was ascertained. A total of 75 (56.8%) isolates were found to harbor at least one of the four urogenes investigated. The maximum number of detected amplicons in one strain was three of the virulence gene regions targeted. Combinations of adhesin and toxin genes encoded by E. coli isolates are presented in Table 3. Considering all virulence genes together, the studied strains exhibited 11 virulence gene profiles, referred to as urovirulence profile (UP) followed by an Arabic numeral. Of these 11 combinations, the most common gene profile was UP1, which was characterized by the presence of only the cnf gene (16 isolates) followed by UP2, which was found in 15 isolates and characterized by the presence of the pap gene only. The least distributed profile was UP11, which was detected in only one strain and characterized by the simultaneous presence of hly and afa genes.

A significant association was detected between the simultaneous presence of hly and pap genes (P< 0.001), which corresponded to 14 (10.6%) strains. Sixteen isolates carried both pap and cnf, 10 strains hly and cnf, and three isolates pap and afa but lacked significance according to the Chi-square and Fisher's tests.
Associations between antimicrobial resistance and virulence traits
Table 4 shows the prevalence of each virulence gene according to antibiotic susceptibility status of isolates. In general, pap was more prevalent in nonsusceptible isolates group (resistant plus intermediately resistant [for FEP: susceptible dose dependent]). Exceptions were beta-lactam antibiotics (IPM, CAZ, FEP, AZT, and CFO), for which susceptible isolates showed a higher prevalence of the pap gene. The afa gene was also more prevalent in nonsusceptible isolates, except TE, AN, FM, and AMC, for which the susceptible isolates showed a higher prevalence. By contrast, the cnf gene was more prevalent in susceptible isolates; the only exceptions were GM and FM for which nonsusceptible isolates showed a higher prevalence. The hly was almost equally distributed between susceptible and nonsusceptible groups although was slightly shifted toward susceptible isolates group and away from nonsusceptible isolates [Table 4].

The possible statistical association between antibiotic susceptibility/nonsusceptibility phenotypes and virulence genes of isolates was subsequently investigated. We found that the distribution of hly and pap in relation to antimicrobial resistance phenotypes was not different (P > 0.05), except that AN-susceptible isolates significantly exhibited a lower prevalence of the hly (P< 0.05). A more analysis revealed two further groups of associations: first, an association between the presence of afa and nonsusceptibility to beta-lactams (CTX and FEP) and the quinolone NA as well as the fluoroquinolone CP (P< 0.05), and second, an association between the presence of cnf and susceptibility to beta-lactams (AM, IPM, CAZ, FEP, AMC), quinolone (NA), and fluoroquinolones (CP, NOR) (P< 0.05) [Figure 1].
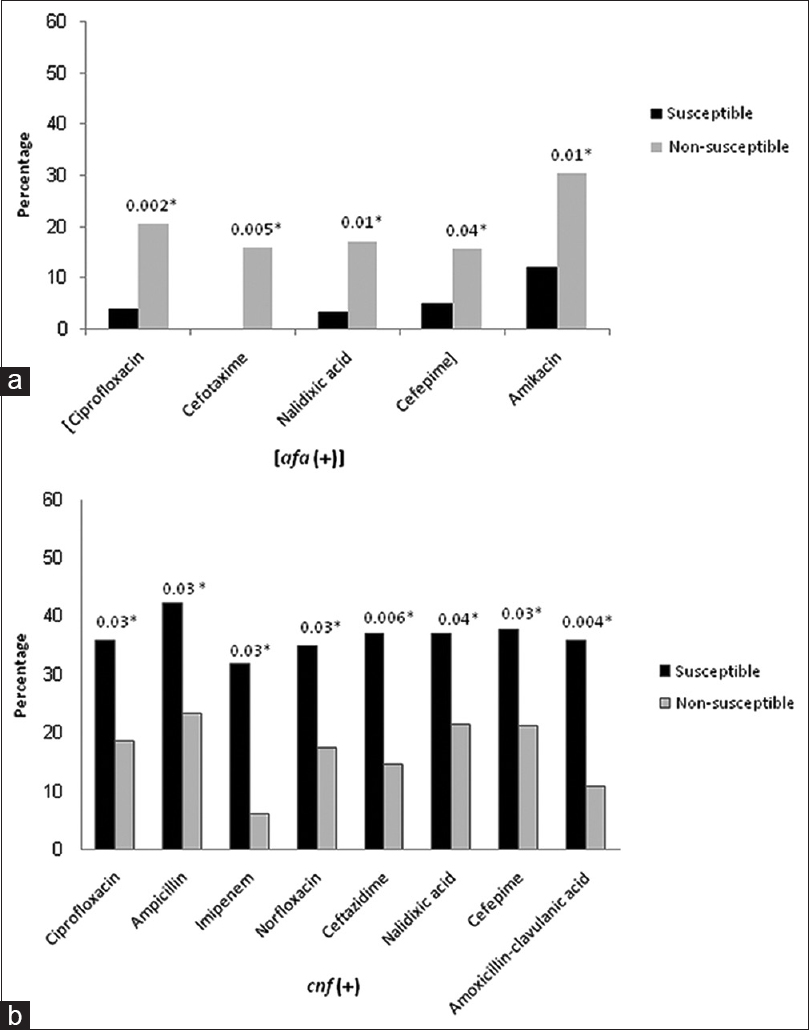
- Relationships between virulence factors and antimicrobial susceptibility: (a) susceptibility to the antibiotics shown was significantly related to the lower prevalence of hly and afa genes. (b) A significant relationship between presence of cnf gene and susceptibility to antibiotics (*P < 0.05)
Discussion
UTI is one of the important bacterial infections, affecting both inpatients and outpatients, and E. coli is the leading causative agent of UTI. It is thought that the pathogenic potential of E. coli isolates is dependent on the presence of various virulence factors.[1] In our study, of the 132 E. coli isolated from UTI, the pap gene was present in 31.1%, the afa in 10.6%, the cnf in 28.8%, and the hly in 16.7% of isolates. The hly was found to be significantly associated with pap. The pap adhesion gene was the most commonly identified virulence gene, in agreement with other reports.[102627] It has been shown that transformation of E. coli with pap sequence makes it a more potent inducer of the host response. It is conceivable that in some strains of E. coli, pap has important role in the progression to more severe infections of the urinary tract.[6] Isolates from outpatients showed a higher prevalence of the studied virulence genes except pap gene.
In recent years, management of UTIs has become increasingly problematic due to the emergence of drug-resistant E. coli strains in many countries.[111328] The emergence of resistance limits the utility of older agents, such as SXT, which results in increased reliance on newer broad-spectrum agents, such as extended-spectrum cephalosporins and fluoroquinolones.[29] Antibiotic sensitivity test in our isolates showed FM as the most effective followed by CFO and IPM. AM, SXT, and CTX were the least effective antibiotics. The isolates from outpatients were more resistant to FM, IPM, AMC, AN, and GM than those from inpatients, in particular AMC. The likely reasons for the high resistance rates to these antibiotics among outpatients are the inappropriate use of these antimicrobials, ineffective infection control and health programs, and cross-resistance among antibiotics of the same class, such as AN and GM.[30] In humans, 80%–90% of antimicrobial drugs are used in outpatients, and it is estimated that 20%–50% of the use of antibiotics is questionable.[31] Resistant strains can be traced from the community to hospitals and contribute to multidrug resistance in hospital settings.[32]
Several studies indicate that resistance to some antibiotics is associated with decreased virulence traits among clinical E. coli isolates.[1516] On the other hand, such phenomenon was not observed in some other studies.[171819] We observed an association between the presence of cnf and susceptibility to the tested beta-lactams, quinolone, and fluoroquinolones [Figure 1]. However, E. coli strains with a reduced susceptibility to the studied extended-spectrum cephalosporins (CTX and FEP), quinolone NA, and fluoroquinolone CP were associated with a significantly increased prevalence of afa. E. coli strains expressing adhesins of the Afa family have unique renal tissue tropisms that favor the establishment of chronic and/or recurrent infections.[8] Moreover, isolates with a reduced susceptibility to AN significantly exhibited a higher prevalence of the hly, resulting in a slightly increased inferred virulence potential compared with susceptible isolates. Compared to their susceptible counterparts, resistant bacterial infections with more virulence factors are generally associated with increased morbidity, mortality, and treatment costs.[33]
The reasons for this correlation are not entirely clear. A biological basis for the association of antimicrobial resistance with virulence genes in E. coli has been previously reported for certain genes; for example, an 80-megadalton plasmid coding for AM resistance has been associated with genes for ST (heat-stable enterotoxin) synthesis.[34] Recently, some in vitro studies have shown that decreased pathogenicity of E. coli is associated with the acquisition of quinolone resistance.[35] Soto et al. suggested that subinhibitory concentrations of quinolones induce the SOS system response (DNA repairing mechanism), which could favor in vitro loss of virulence genes in E. coli strains. According to their results, the virulence genes may have been lost in exchange for resistance.[36] However, the finding that spontaneous fluoroquinolone-resistant mutants derived from hemolytic fluoroquinolone-susceptible strains are still able to produce hemolysin[18] suggested otherwise. Although one hypothesis does not exclude the other, further studies are needed to consolidate the findings. It is also possible that ecological factors such as the geographic origin of isolates represent important additional factors that should be considered.[37] It is worthwhile to investigate whether gene linkage on plasmids or other mobile genetic elements underlies the associations observed in our study.
In conclusion, this study describes the prevalence of resistance phenotypes and virulence genes in E. coli isolates from UTI in Kurdistan Province, Iran. The study also shows the relationship between antimicrobial resistance and virulence genes. The increasing emergence of antimicrobial resistance and relationships between resistance and virulence genes suggest that there is a great need for surveillance programs to monitor drug resistance in pathogenic bacteria. Such surveillance programs would provide appropriate guidelines for restriction of antimicrobial use and would be important steps in efforts to understand, prevent, and control the emergence and spread of antimicrobial resistance and virulence genes.
Financial support and sponsorship
This study was supported by Faculty of Medicine, Kurdistan University of Medical Sciences, Sanandaj, Iran.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We are grateful to the staff of the microbiology laboratory at Besat and Tohid Hospitals for collecting E. coli isolates used in this study.
References
- Relationship between susceptibility to antimicrobials and virulence factors in paediatric Escherichia coli isolates. Int J Antimicrob Agents. 2008;31(Suppl 1):S4-8.
- [Google Scholar]
- Virulence genes, quinolone and fluoroquinolone resistance, and phylogenetic background of uropathogenic Escherichia coli strains isolated in Japan. Jpn J Infect Dis. 2010;63:113-5.
- [Google Scholar]
- Recurrent urinary tract infections management in women: A review. Sultan Qaboos Univ Med J. 2013;13:359-67.
- [Google Scholar]
- Integrons in uropathogenic Escherichia coli and their relationship with phylogeny and virulence. Microb Pathog. 2014;77:73-7.
- [Google Scholar]
- Uropathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013;12:8.
- [Google Scholar]
- Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. J Microbiol Immunol Infect. 2014;47:455-61.
- [Google Scholar]
- Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62:84-90.
- [Google Scholar]
- Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. 2012;2012:681473.
- [Google Scholar]
- Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One. 2011;6:e18063.
- [Google Scholar]
- Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17:e450-3.
- [Google Scholar]
- Rapid emergence of ciprofloxacin-resistant Enterobacteriaceae containing multiple gentamicin resistance-associated integrons in a Dutch hospital. Emerg Infect Dis. 2001;7:862-71.
- [Google Scholar]
- Broad resistance due to plasmid-mediated AmpC beta-lactamases in clinical isolates of Escherichia coli. Clin Infect Dis. 2002;35:140-5.
- [Google Scholar]
- Co-trimoxazole and quinolone resistance in Escherichia coli isolated from urinary tract infections over the last 10 years. Int J Antimicrob Agents. 2005;26:75-7.
- [Google Scholar]
- Association between antimicrobial resistance and virulence in Escherichia coli. Virulence. 2012;3:18-28.
- [Google Scholar]
- Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J Infect Dis. 2003;188:759-68.
- [Google Scholar]
- Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother. 2006;57:204-11.
- [Google Scholar]
- Virulence genotype and phylogenetic origin in relation to antibiotic resistance profile among Escherichia coli urine sample isolates from Israeli women with acute uncomplicated cystitis. Antimicrob Agents Chemother. 2005;49:26-31.
- [Google Scholar]
- Relationship between haemolysis production and resistance to fluoroquinolones among clinical isolates of Escherichia coli. J Antimicrob Chemother. 1999;43:277-9.
- [Google Scholar]
- Phylogenetic background and carriage of pathogenicity island-like domains in relation to antibiotic resistance profiles among Escherichia coli urosepsis isolates. J Antimicrob Chemother. 2006;58:748-51.
- [Google Scholar]
- Guidelines on urological infections. 2015. European Association of Urology. Available from: http://uroweb.org/wp-content/uploads/19-Urological-infections_LR2.pdf
- [Google Scholar]
- Biochemical and molecular characterization of Cronobacter spp. (formerly Enterobacter sakazakii) isolated from foods. Antonie Van Leeuwenhoek. 2011;99:257-69.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing, 24th Informational Supplement (M100.S24) Wayne Pa: Clinical and Laboratory Standards Institute; 2014. p. :34.
- [Google Scholar]
- Genotyping and characterization of CTX-M-15 -producing Klebsiella pneumoniae isolated from an Iranian hospital. J Chemother. 2016;28:289-96.
- [Google Scholar]
- Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12:85-90.
- [Google Scholar]
- Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol. 1992;30:1189-93.
- [Google Scholar]
- Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev Inst Med Trop Sao Paulo. 2008;50:255-60.
- [Google Scholar]
- The association of virulence determinants of uropathogenic Escherichia coli with antibiotic resistance. Jundishapur J Microbiol. 2014;7:e9936.
- [Google Scholar]
- Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011;10:4114-25.
- [Google Scholar]
- Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med. 2001;135:41-50.
- [Google Scholar]
- Community factors in the development of antibiotic resistance. Annu Rev Public Health. 2007;28:435-47.
- [Google Scholar]
- The use and resistance to antibiotics in the community. Int J Antimicrob Agents. 2003;21:297-307.
- [Google Scholar]
- Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J Infect Dis. 2002;186:49-56.
- [Google Scholar]
- Excess infections due to antimicrobial resistance: The “Attributable fraction”. Clin Infect Dis. 2002;34(Suppl 3):S126-30.
- [Google Scholar]
- Linkage of heat-stable enterotoxin activity and ampicillin resistance in a plasmid isolated from an Escherichia coli strain of human origin. Infect Immun. 1980;30:617-20.
- [Google Scholar]
- Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis. 2002;186:1039-42.
- [Google Scholar]
- Quinolones induce partial or total loss of pathogenicity islands in uropathogenic Escherichia coli by SOS-dependent or – Independent pathways, respectively. Antimicrob Agents Chemother. 2006;50:649-53.
- [Google Scholar]
- Antimicrobial resistance, virulence genes, and phylogenetic background in Escherichia coli isolates from diseased pigs. FEMS Microbiol Lett. 2010;306:15-21.
- [Google Scholar]







