Translate this page into:
Etiological Spectrum of Infective Diarrhea in Renal Transplant Patient by Stool PCR: An Indian Perspective
Address for correspondence: Dr. Vaibhav Tiwari, Department of Nephrology, Sir Ganga Ram Hospital, Old Rajinder Nagar, New Delhi - 110 060, India. E-mail: drvt87@gmail.com
-
Received: ,
Accepted: ,
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
This article was originally published by Wolters Kluwer - Medknow and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction:
Diarrhea is a common cause of morbidity and mortality among renal transplant patients. The etiological spectrum of pathogens varies with regional diversity, socioeconomic conditions, sanitation, and eating habits. We aimed to delineate the etiological profile of gastrointestinal pathogens in renal transplant patients using the stool Polymerase chain reaction.
Methods:
In this single-center, retrospective analysis of patients from January 2016 to January 2018, all renal transplant patients who were admitted with severe diarrhea and underwent the stool Polymerase chain reaction (PCR) were included. In the control group, we included patients from the general population who were admitted with similar complaints in the general medicine ward and underwent stool PCR over the same duration.
Results:
One hundred ten admissions occurred over 2 years in the transplant group. 86% of samples were positive for infection. More than one organism was seen in 68% of the patient. Norovirus was the most common organism isolated. Giardia lamblia with Norovirus was the most common coinfection among the transplant population. In the control group, 87% of samples tested positive, with 53% of patients having more than one organism. Enteroaggregative E. coli was the common organism, Enteroaggregative E. coli with Enteropathogenic E. coli and Enterotoxigenic E. coli were the most common organism in combination. Both the groups had similar incidence of infection with multiple organisms.
Conclusion:
The etiological profile of gastrointestinal pathogens differs significantly between the transplant and general population. Coinfections are common in both populations. Norovirus is the most common pathogen in the transplant population, presenting as isolated as well as in coinfections.
Keywords
Diarrhea
Infective diarrhea
renal transplant
stool PCR
Introduction
Acute and chronic diarrhea is every transplant physician's nightmare. The incidence of diarrhea in transplant patients ranges from 20-50%.[123] From adjusting immunosuppression to searching the etiology is a drill that has to be played every single time patient comes to a physician's clinic. Diarrhea in transplant patients differs in etiology ranging from infection to drug side effect, consequences ranging from AKI to graft rejection. There is a wide variation in infective etiology among patients that is mainly based on geographical factors, socio-economic status, eating habits, and access to clean water. Most of the studies in the transplant for evaluation of diarrhea were conducted in western countries. Because India, with its poor socio-economic conditions, differs from developed western countries, so does the etiological profile of infective organism causing diarrhea. This study was aimed to evaluate the etiological spectrum of organisms causing diarrhea in transplant patients and compare it with the general population with similar complaints with the help of the stool Polymerase chain reaction (PCR) test.
Materials and Methods
We planned a retrospective cohort study of renal transplant recipients with concurrent non-transplant controls. For cases, we reviewed records of all renal transplant recipients >18 years of age, admitted in our hospital with complaints of diarrhea (>3 bowel movements of liquid consistency for more than three days and less than 14 days) from January 2016 to January 2018. Only those who underwent a stool PCR examination were included. Exclusion criteria were patients on chemotherapy, malabsorption syndrome, inflammatory bowel disease, irritable bowel syndrome, and patients with known causes of non-infective diarrhea.
For the control group, all patients more than 18 years of age admitted in the medicine unit with the complaint of diarrhea and who underwent stool PCR examination during the same period were included. Inclusion and exclusion criteria were same as of the transplant group.
Multiplex PCR
The FilmArray GI panel test is FDA, Conformité Européene–In Vitro Diagnostics (CE-IVD) and Therapeutic Goods Administration (TGA) certified nested PCR assay. Two hundred microliters of the specimen in Cary-Blair transport medium was subject to FilmArray GI Panel testing, according to the manufacturer's instructions. The FilmArray GI Panel test consists of automated nucleic acid extraction, reverse transcription, amplification, and analysis, with results available in 1 h per run per specimen. Each FilmArray GI Panel pouch contains an internal nucleic acid extraction control and PCR control. The FilmArray GI Panel runs were considered valid if the run completed normally and internal controls passed. The FilmArray GI Panel software performs automated result analysis with each target in a valid run reported as detected or not detected. If either internal control fails, the software automatically provides a result of invalid for all panel analytes. All specimens having invalid results were retested.
The panel targets include:-
| Bacteria | Campylobacter (jejuni, coli, and upsaliensis), Clostridium difficile [toxins A/B], Plesiomonas shigelloides, Salmonella, Yersinia enterocolitica, non-cholera Vibrio [parahaemolyticus, and vulnificus], Vibrio cholera, E. coli O157, Enteroaggregative E. coli [EAEC], Enteropathogenic E. coli [EPEC], Enterotoxigenic E. coli [ETEC], Shigalike toxin-producing E. coli [STEC], Shigella/Enteroinvasive E. coli [EIEC] |
| Virus | Adenovirus F40/41, Astrovirus, Norovirus GI/GII, Rotavirus A, Sapovirus [I, II, IV, and V] |
| Parasites | Cryptosporidium spp, Cyclospora cayetanensis, Entamoeba histolytica, Giardia lamblia |
CMV
All transplant patients with diarrhea but negative stool PCR, CMV RT-PCR, and colonoscopy were done. Both the tests were also sought when the patient presented with >2 organ involvement along with diarrhea. Test to diagnose CMV infections were not done in control group
Real-time PCR for CMV DNA quantification
QPCR for CMV was performed in both biopsy and plasma samples in all the patients using the artus CMV RG PCR kit (Qiagen, Hilden, Germany), as per the manufacturer's instructions. The assay targets a 105 bp region of the glycoprotein gene of CMV genome. Internal control was added to each sample before extraction, and each batch of PCR included four positive and one negative control. CMV viral load levels were then expressed as the number of CMV DNA copies/mg of tissue for intestinal biopsy samples and in copies/mL for plasma samples. The dynamic range of the assay is 102–106 copies/mL with a lower limit of detection of 57.1 copies/mL.
Extraction of DNA from intestinal biopsy and blood
All the biopsy samples were collected in a sterile plain vial and transported to the laboratory on ice. Extraction of DNA from biopsy tissue sample was done using the automated DNA extraction system (QIAsymphony, Qiagen, Germany) using the DSP virus/pathogen kit, with slight modifications. Tissue was weighed, and 5 mg of the tissue was digested by proteinase K before processing. The volume was adjusted to 750 μl with AVE buffer (Qiagen, Germany). Plasma (750 μl) samples were directly processed in the automated Qiasymphony, as per manufacturer instructions.
Statistical analysis
Quantitative values for variables were given as medians ± extreme values or means ± standard deviations, and qualitative data were given as numbers and percentages. Pearson's Chi-square test or the Fisher exact test was used to compare qualitative variables. Between-group comparison of numeric parametric data was done by unpaired t-test. Results were considered to be statistically significant for two-sided P values < 0.05.
Results
A total of 130 renal transplant recipients were admitted with a chief complaint of diarrhea during the study period. All consecutive patients could not be included as only 105 patients (81%) underwent a Stool Multiplex PCR study. These patients were included for 110 diarrheal events, defined as hospital admission for diarrhea. During the same period, 350 patients nontransplant patients were admitted with diarrhea. Stool PCR was done for 188 patients (30% of the patients were excluded due to non-availability of Stool PCR study) for 194 diarrheal events and were included in the control group for analysis.
Baseline characteristics of patients have been shown in Table 1. The transplant group had higher numbers of males and diabetics as compared to the control group. Baseline serum creatinine was significantly higher in the transplant group (1.43 vs. 0.98; P value = 0.02). The mean duration of the transplant was three years; 15% of patients had a transplant within six months. The duration of diarrhea was comparable in both the groups (six days vs. four days; P value 0.79). More than 90% of patients in the transplant group had ATG as induction agent with Tacrolimus, MMF, and prednisolone as current immunosuppression. The frequency of stools was higher in the control group, although it was not significant. Abdominal pain was the most common symptom in both groups [Table 1]. The incidence of fever, along with diarrhea, was similar in both the groups. History of intake of food from outside home was present in nearly 50% of patients in both groups.
| Baseline characteristics | Transplant group | Control group | P |
|---|---|---|---|
| Age, years | 46±12.33 | 41±14.65 | NS |
| Sex, (Male%) | 79% | 58% | 0.03 |
| Diabetes (%) | 65% | 23% | 0.01 |
| Duration of diarrhea before admission, days | 7.2±3.4 | 4.5±2.3 | NS |
| Frequency of Stools/day | 6.6±3.1 | 7.3±2.5 | NS |
| Symptoms,% | NS | ||
| Abdominal pain | 60 | 72 | NS |
| Fever | 17 | 10 | NS |
| Vomiting | 19 | 12 | NS |
| Loss of appetite | 25 | 11 | NS |
| History of outside food consumption | 47% | 52% | NS |
| Laboratory parameter | |||
| Serum creatinine | 1.43±1.31 | 0.98±1.01 | 0.02 |
| Transplant characteristics | |||
| Duration of Transplantation, years | 3.1±2.1 | - | |
| Induction Agent | |||
| ATG | 94% | - | |
| Basilixmab | 4% | - | |
| No Induction | 2% | - | |
| Immunosuppresive drugs | |||
| Tac + MMF + Pred | 85% | - | |
| Tac + Aza + Pred | 9% | - | |
| Cyclo + MMF + Pred | 4% | - | |
| Cyclo + Aza + Pred | 2% | - |
NS - Not Significant, Tac - Tacrolimus, MMF - Mycofenolate Mofetil, Aza - Azathioprine, Pred - Prednisolone. ATG - Antithymocyte globulin
Microbiological yield
A total of 86.3% of samples yielded one or more organisms in stool PCR. A total of 181 organisms were isolated from 110 diarrheal event. Bacterial infection was the most common etiology, with almost 50% of all infections being bacterial in origin [Figure 1]. The majority of infections were coinfections, with 68% of positive PCR had two or more organisms [Figure 2]. Overall, Norovirus G1/G2 (20%) was the most common organism isolated, followed by G. lamblia (17%), and Enteropathogenic Escherichia coli (EPEC) (16%) [Table 2]. The frequency of Norovirus infection was significantly higher as overall infection as well as isolated infection in the transplant group than the control group [Table 3]. Frequency of Cryptosporidium spp. and EPEC was also significantly higher in the transplant group than the control group. PCR was positive in 87.4% of samples in the control group. 270 organisms were isolated from the 194 diarrheal events. GI Infection was dominated by bacteria 64.4%, followed by an equal incidence of parasite and viral infection [Figure 1]. Overall, G. lamblia, Enteroaggregative E. coli (EAEC), Shigella/Enterotoxigenic E. coli (ETEC) were present in equal frequency (13%) [Tables 2 and 3]. Clostridium difficile, Vibrio cholerae, Salmonella, and Rotavirus were significantly higher in the control group than the transplant group.

- Graph showing the frequency of various categories of gastrointestinal pathogens detected by stool PCR in each group
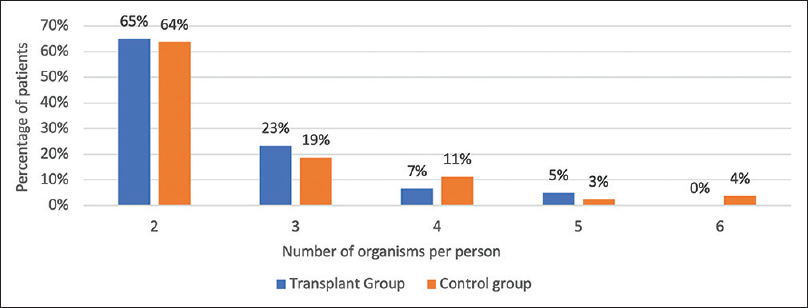
- Graph showing the frequency of the number of organisms per person in coinfection detected by stool PCR in each group
| Organisms | Transplant group | Control group | P |
|---|---|---|---|
| Norovirus GI/II | 37 (20%) | 23 (9%) | 0.004 |
| Giardia lamblia | 30 (17%) | 35 (13%) | NS |
| Enteropathogenic Escherichia coli (EPEC) | 29 (16%) | 22 (8%) | 0.015 |
| Cryptosporidium spp. | 21 (12%) | 10 (4%) | 0.002 |
| Shigella/Enteroinvasive E. coli (EIEC) | 20 (11%) | 34 (13%) | NS |
| Enteroaggregative E. coli (EAEC) | 17 (9%) | 35 (13%) | NS |
| Enterotoxigenic E. coli (ETEC) | 12 (7%) | 18 (7%) | NS |
| Campylobacter spp. | 7 (4%) | 22 (8%) | 0.07 |
| Clostridium difficile | 3 (2%) | 16 (6%) | 0.03 |
| Vibrio cholerae | 3 (2%) | 17 (6%) | 0.02 |
| Plesiomonas shigelloides | 1 (1%) | 1 (0%) | NS |
| Sapovirus | 1 (1%) | 6 (2%) | NS |
| Salmonella spp. | 0 (0%) | 9 (3%) | 0.013 |
| E. histolytica | 0 (0%) | 3 (1%) | NS |
| Cyclospora | 0 (0%) | 2 (1%) | NS |
| Adenovirus | 0 (0%) | 2 (1%) | NS |
| Rotavirus | 0 (0%) | 10 (4%) | 0.007 |
| Astrovirus | 0 (0%) | 5 (2%) | 0.09 |
NS - Not Significant
| Organisms | Transplant group | Control group | P |
|---|---|---|---|
| Norovirus GI/II | 8 (27%) | 6 (8%) | 0.02 |
| G. lamblia | 7 (23%) | 8 (10%) | NS |
| Campylobacter spp. | 5 (17%) | 5 (6%) | NS |
| Shigella/Enteroinvasive E. coli (EIEC) | 5 (17%) | 10 (13%) | NS |
| Enteropathogenic Escherichia coli (EPEC) | 3 (10%) | 2 (3%) | NS |
| Enteroaggregative E. coli (EAEC) | 1 (3%) | 10 (13%) | NS |
| Clostridium difficile (Toxin A/B) | 1 (3%) | 8 (10%) | NS |
| Cryptosporidium spp. | 1 (3%) | 5 (6%) | NS |
| Salmonella spp. | 0 (0%) | 2 (2%) | NS |
| V. cholerae | 0 (0%) | 9 (11%) | NS |
| Enterotoxigenic E. coli (ETEC) | 0 (0%) | 3 (4%) | NS |
| Rotavirus | 0 (0%) | 4 (5%) | NS |
| Sapovirus | 0 (0%) | 3 (4%) | NS |
| Astrovirus | 0 (0%) | 3 (4%) | NS |
| E. histolytica | 0 (0%) | 2 (3%) | NS |
NS - Not Significant
Coinfections diagnosed by Stool PCR
In the transplant group, 151 (83%) organisms were found in various combinations in 65 (68%) diarrheal events. 37% of diarrheal events had three or more organisms detected in the stool [Figure 2]. G. lamblia with Norovirus (15%) was the most common infection followed by G. lamblia with Cryptosporidium spp. (13%) [Table 4]. In the control population, 80 diarrheal events (53%) had multiple organisms detected in the single diarrheal event. Twenty-nine patients (36%) had three or more organisms in the stool. EAEC with EPEC (18%) was the most common combination followed by EAEC with ETEC (11%) and EPEC with ETEC (11%). Coinfection of Cryptosporidium spp. and G. lamblia was significantly higher in the transplant group.
| Organismsd | Transplant group | Control group | P |
|---|---|---|---|
| G. lamblia + Norovirus | 13 (15%) | 4 (6%) | NS |
| Cryptospridium spp. + G. lamblia | 11 (13%) | 2 (3%) | 0.04 |
| Cryptospridium spp. + Norovirus | 10 (11%) | 2 (3%) | 0.07 |
| EAEC + EPEC | 9 (10%) | 12 (18%) | NS |
| EPEC + Norovirus | 9 (10%) | 4 (6%) | NS |
| EPEC + ETEC | 8 (9%) | 7 (11%) | NS |
| G. lamblia + EIEC | 7 (8%) | 4 (6%) | NS |
| EAEC + G. lamblia | 6 (7%) | 3 (5%) | NS |
| EPEC + G. lamblia | 4 (5%) | 5 (8%) | NS |
| G. lamblia + ETEC | 4 (5%) | 3 (5%) | NS |
| EAEC + ETEC | 3 (3%) | 7 (11%) | NS |
| EPEC + EIEC | 3 (3%) | 4 (6%) | NS |
| EIEC + ETEC | 1 (1%) | 4 (6%) | NS |
| EAEC + EIEC | 0 (0%) | 5 (8%) | 0.013 |
NS - Not Significant
Twenty transplant patients who had stool PCR negative, underwent RT-PCR for CMV. None of them had positive results. Five patients also underwent colonoscopy and biopsy. None of them had pathological findings supportive of CMV disease.
Discussion
This is a first report evaluating the infective etiology of diarrheal illness among renal transplants by multiplex PCR from the Indian subcontinent. PCR is a state-of-the-art technology which requires less time, human resources, and provides more diagnostic accuracy. In a study by Coste JF et al. in 49 solid organ transplant (SOT) patients, PCR was compared with conventional methods. Conventional methods had a poor diagnostic performance with a detection rate of less than one third as compared to stool PCR (23% vs. 72%).[4] In the study by Amar CFL et al., the fecal samples of the original Infectious Intestinal Disease (IID) study performed on more than 4000 non-transplant patients in the UK were reassessed by Stool PCR and results were compared with the original study which has used traditional methods including stool microscopy and culture.[56] In the cases, after the application of PCR, the rate of detection at least a single organism was increased to 75% from 50%. Norovirus was the most common enteric pathogen (36%) instead of Campylobacter jejuni (9%), which was found to be most frequent in IID. In the control population, the detection rate increased to 42% from 19%. The American Society of Transplantation Infectious Diseases Community of Practice now recommends performing stool PCR in SOT patients presenting with diarrhea.[7] PCR has several limitations in clinical practice. It is an expensive test as compared to the conventional methods. It does not distinguish between acute and asymptomatic carrier state. It also does not provide a drug sensitivity pattern for guiding the clinician. Nevertheless, in transplant patients, where stakes are high, delay in diagnosis may result in unnecessary modification in immunosuppressive medicines and ordering unnecessary tests. Stool PCR provides results in few hours, detecting multiple organisms at once, thus enabling the clinician to make a quick decision, avoid unwanted complications.
The spectrum of diarrhea in the non-transplant population
The spectrum of etiological agents was much more diverse among the control population. A comparison of various studies regarding the frequency of the infective organisms has been shown in Table 5. Most of the studies were conducted in west and tested only few organisms.[58910] Findings in our study closely matches with the study from Turkey by Arslan et al., although they had a small sample size (n = 33).[8] More than 85% of cases were positive for an infectious agent; Giardia lamblia was the most common organism isolated in both studies. In other study from south India by Ramakrishna B et al., using stool PCR, evaluated 106 patients with acute and chronic diarrhea.[11] 50% of the sample yielded one or more organisms. As compared to our findings of 53% coinfection among controls, in their study, 48% had coinfections. Similar to our results, EAEC was the most organisms, EAEC and EPEC and EAEC and ETEC were the most common coinfections. This reinforces the fact that the spectrum of infective gastrointestinal illness largely depends upon the region, food habits, and socio-economic state of the population.
| Study | Beal SG et al.[9] | Amar CFL et al.[5] | Halligan et al.[10] | Arsalan et al.[8] | Present study |
|---|---|---|---|---|---|
| Year | 2018 | 2007 | 2013 | 2007 | 2019 |
| Population | Adult | Adult+children | adult | Adults | Adults |
| Region | USA | UK | UK | Turkey | India |
| Number | 180 | 2422 | 262 | 33 | 194 |
| positivity | 15.60% | 53% | 22.10% | 89% | 87.40% |
| Organisms, overall | |||||
| Norovirus GI/II | 8.30% | 36% | 9.10% | - | 9% |
| Giardia lamblia | 1.10% | 2% | 1.4% | 21.20% | 13% |
| Enteropathogenic Escherichia coli (EPEC) | 11.00% | - | - | - | 8% |
| Cryptosporidium spp. | 0.60% | 2% | 0.60% | 3% | 4% |
| Shigella/Enteroinvasive E. coli (EIEC) | 1.10% | - | 1.30% | - | 13% |
| Enteroaggregative E. coli (EAEC) | 1.10% | 6% | - | - | 13% |
| Enterotoxigenic E. coli (ETEC) | 0.60% | - | - | - | 7% |
| Campylobacter spp. | 0.60% | 23% | 5.60% | 6% | 8% |
| Clostridium difficile | - | - | 5.80% | 6% | 6% |
| Vibrio cholerae | 1.10% | - | - | - | 6% |
| Plesiomonas shigelloides | 0.60% | - | - | - | 0% |
| Sapovirus | 2.80% | 4% | - | - | 2% |
| Salmonella spp. | 0.60% | 6% | 3% | 12.10% | 3% |
| E. histolytica | 0% | - | 0.60% | 21.20% | 1% |
| Cyclospora | 0.60% | - | - | - | 1% |
| Adenovirus | 0.60% | - | 0.80% | 3% | 1% |
| Rotavirus | 3.90% | 31% | 2.70% | 6% | 4% |
| Astrovirus | 1.10% | - | - | - | 2% |
| E. coli O 157 | - | - | 0.30% | - | 0% |
| CMV | - | - | - | 0 | - |
| Shigalike toxin-producing E. coli (STEC) | - | - | - | - | 0% |
| Blastocystis hominis | - | - | - | 9% | - |
Prevalence and Spectrum of infective diarrhea in the transplant population
The prevalence of infective diarrhea among the transplant recipient has been varied from 38-77% depending upon the region and method of diagnosis.[8121314] In the landmark trial, the DIDACT study, 69 of 108 (64%) patients with diarrhea had infective origin.[15] In a study from turkey, renal and liver transplant recipients were compared with the immunocompetent population for the cause of diarrhea.[8] Infective cause for diarrhea was elucidated in 76% of transplant patients and 89% of the immunocompetent population. In a study from America, liver and kidney transplant recipients were compared with community-acquired and hospital-acquired diarrhea.[16] The majority of episodes had no identifiable cause (60-75%) and were self-limited (90%). The infective cause was found in 30%, and 19% of community-acquired and hospital-acquired diarrhea, respectively.
The etiological spectrum of infective agents in acute gastroenteritis in transplant patients is mainly region centered. Table 6 reviews and compares the various pathogenic organisms found in recent studies in the transplant population. In the present study, bacterial were the most common among all organisms in both cases and control, although individually Norovirus and G. lamblia were the most common among cases and controls, respectively.
| Author | Altiparmark MR et al.[12] | Maes B et al.[15] | Coste JFL et al.[4] | Arsalan et al.[8] | Present study |
|---|---|---|---|---|---|
| Year | 2002 | 2006 | 2013 | 2007 | 2019 |
| Population | Renal transplant recipients | Renal transplant recipients | Renal transplant recipients | Solid organ transplant recipients | Renal transplant recipients |
| Region | Turkey | Belgium | France | Turkey | India |
| Number of stool sample tested | 41 | 108 | 54 | 33 | 110 |
| % of Positives | 41.5%% | 20% | 72% | 82.60% | 86.30% |
| Organisms, overall | |||||
| Norovirus GI/II | - | - | 36% | - | 20% |
| Giardia lamblia | 18% | - | 2% | 27.20% | 17% |
| Enteropathogenic Escherichia coli (EPEC) | - | - | 38% | - | 16% |
| Cryptosporidium spp. | - | - | 2% | 21.20% | 12% |
| Shigella/Enteroinvasive E. coli (EIEC) | 18% | - | - | - | 11% |
| Enteroaggregative E. coli (EAEC) | - | - | - | - | 9% |
| Enterotoxigenic E. coli (ETEC) | - | - | - | - | 7% |
| Campylobacter spp. | - | 65% | 38% | 6% | 4% |
| Clostridium difficile | 12% | - | 2% | 9% | 2% |
| Vibrio cholerae | - | - | - | - | 2% |
| Plesiomonas shigelloides | - | - | - | - | 1% |
| Sapovirus | - | - | - | - | 1% |
| Salmonella spp. | - | 9% | 2% | 3% | 0% |
| E. histolytica | 6% | - | - | 3% | 0% |
| Cyclospora | - | - | - | - | 0% |
| Adenovirus | - | - | - | - | 0% |
| Rotavirus | - | - | 4% | 3% | 0% |
| Astrovirus | - | - | 2% | - | 0% |
| CMV | 12% | - | - | 18% | 0% |
| Shigalike toxin-producing E. coli (STEC) | - | - | 6% | - | 0% |
| Hymenolepsis nana | 6% | - | - | - | - |
| Blastocystis hominis | 12% | - | - | 3% | - |
| Candida albicans | 18% | - | - | - | - |
| Shigella sonnei | - | - | - | 9% | - |
Co-infections and its implication in the transplant population
Co-infection is a common finding in an acute diarrheal illness.[1718192021] Stool PCR is superior in detecting the multiple organisms than the conventional methods.[45] In the study by Coste et al., conventional methods failed to detect any co-infection in the transplant population, whereas the stool PCR found that 23% infections were actually co-infections.[4] In the study, Amar et al., after the application of PCR, the detection rate of multiple infections significantly increased by 73%.[5] Many of the studies have looked upon a specific association between viral and bacterial infections.[172223] In this study, co-infections were more in the transplant group (67% vs. 57%), but it did not reach the statistically significant level (p = 0.06). Existence of gut flora as commensal is a known fact but do the gut pathogens have a symbiotic, independent or suppresses each other's growth is largely unknown. According to a study by Byoyofo et al., parasites have specifically lower prevalence with viral infections. This was hypothesized due to antiviral cytokines like interferon are upregulated locally by the parasite, the viral infection is suppressed in the presence of parasitic infection.[22] Paradoxically, in this study, the most common combination was found to be with Norovirus with G. lamblia (19%) in the transplant group. This may be attributed to the immunosuppressed state of the transplant recipients. In the controls, we did get fewer patients who have co-infections that include viruses with the parasites. Many studies have correlated the severity of diarrhea in terms of duration and number of stools/days with the co-infections.[2425262728] In our study, no such pattern was observed, and clinical features were inadequate to predict the multiplicity of organisms and vice-versa. One of the potential benefits of diagnosing coinfections apart from more targeted therapy is the prevention of incompletely diagnosed case who could be either termed as resistant infection or attributing it to immunosuppression and subsequently modification of the drugs.
Norovirus was the most common infective pathology in 20% of transplant patients. Norovirus is the leading cause of diarrhea in the united states, even among non-transplant patient.[29] Norovirus is notorious for causing chronic infection among immunocompromised patients.[30313233] In a study by Roos et al., they demonstrated Sapo/Norovirus was present in 90% of patients with chronic diarrhea.[34] Chronic Norovirus is a known complication in renal transplant patients.[35] The incidence of Norovirus from India in the transplant population is mostly lacking. The prevalence of Norovirus in India is 5–15% in children presenting with acute gastroenteritis.[363738] In our control population, 9% of patients were harboring Norovirus at the time of presentation. Although the incidence is almost half of the transplant population, nevertheless, Norovirus thus contributes to a chunk of cases among the general population, which mostly go undiagnosed as conservative methods do not detect the virus.
G. lamblia is endemic in India, and its prevalence range from 5.5% to 70%, especially in children from northern India in community-based surveys.[39] Intestinal parasite infestation is generally rare among developed countries, attributing it to better socio-economic conditions. It is the second most common intestinal pathogens detected in our study (17%). Giardiasis also has frequent occurrence along with other pathogens in transplant patients.
The incidence of diarrhea due to CMV disease is estimated to be 1-20%.[81215164041] In the present study, we did not find any CMV cases in patients presented over two years. Since only 18% of patients underwent CMV PCR, and only 4% underwent colonoscopy, there could be a chance of under-diagnosis of the infection. Further studies that are specifically designed to detect CMV are needed to get the actual incidence of the disease.
There are several limitations to the study. First, due to its retrospective nature, complete elimination of selection and information bias was not possible. Second, rather than including all consecutive patients of diarrhea, only those patients were included who were admitted and underwent stool PCR, this have resulted in missing a considerable number of cases (29%). As such the results in this study provide a glimpse of problem of post-transplant diarrhea and further studies are needed to provide a complete picture of this issue.
Conclusion
The transplant population has a different infective pathogen spectrum than the general population. Co-infections are common in transplants as well as the general population. Norovirus is the most common organism in the transplant group as an isolated infection as well as in coinfections. G. lamblia and E. Coli spp. are more common in the general population.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Gastrointestinal complications in renal transplant recipients: MITOS Study. Transplant Proc. 2007;39:2190-3.
- [Google Scholar]
- Gastrointestinal complications in liver transplant recipients: MITOS Study. Transplant Proc. 2007;39:2311-3.
- [Google Scholar]
- Association between gastrointestinal symptoms and health-related quality of life after heart transplantation. J Heart Lung Transplant. 2010;29:1388-94.
- [Google Scholar]
- Microbiological diagnosis of severe diarrhea in kidney transplant recipients by use of multiplex PCR assays. J Clin Microbiol. 2013;51:1841-9.
- [Google Scholar]
- Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: Re-examination of the English case-control Infectious Intestinal Disease Study (1993-1996) Eur J Clin Microbiol Infect Dis. 2007;26:311-23.
- [Google Scholar]
- A study of infectious intestinal disease in England: Microbiological findings in cases and controls. Commun Dis Public Health. 1999;2:108-13.
- [Google Scholar]
- Diagnosis and management of diarrhea in solid-organ transplant recipients: Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant. 2019;33:e13550.
- [Google Scholar]
- Etiologic agents of diarrhea in solid organ recipients. Transpl Infect Dis. 2007;9:270-5.
- [Google Scholar]
- A gastrointestinal PCR panel improves clinical management and lowers health care costs. J Clin Microbiol. 2018;56:e01457-17.
- [Google Scholar]
- Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: A comparative diagnostic and clinical utility study. Clin Microbiol Infect. 2014;20:O460-7.
- [Google Scholar]
- Utility of multiplex polymerase chain reaction (PCR) in diarrhea—An Indian perspective. Indian J Gastroenterol. 2018;37:402-9.
- [Google Scholar]
- Cryptosporidiosis in paediatric renal transplantation. Pediatr Nephrol. 2009;24:2245-55.
- [Google Scholar]
- Infectious gastroenteritis in bone-marrow-transplant recipients. N Engl J Med. 1982;306:1010-2.
- [Google Scholar]
- Severe diarrhea in renal transplant patients: Results of the DIDACT Study. Am J Transplant. 2006;6:1466-72.
- [Google Scholar]
- Diagnostic yields in solid organ transplant recipients admitted with diarrhea. Clin Infect Dis. 2015;60:729-37.
- [Google Scholar]
- Clinical features of acute gastroenteritis associated with rotavirus, enteric adenoviruses, and bacteria. Arch Dis Child. 1986;61:732-8.
- [Google Scholar]
- Mixed viral-bacterial infections and their effects on gut microbiota and clinical illnesses in children. Sci Rep. 2019;9:865.
- [Google Scholar]
- Comprehensive analysis of prevalence, epidemiologic characteristics, and clinical characteristics of monoinfection and coinfection in diarrheal diseases in children in Tanzania. Am J Epidemiol. 2017;186:1074-83.
- [Google Scholar]
- Etiology of acute gastroenteritis in hospitalized children in Melbourne, Australia, from April 1980 to March 1993. J Clin Microbiol. 1998;36:133-8.
- [Google Scholar]
- Multiple etiologies of infectious diarrhea and concurrent infections in a pediatric outpatient-based screening study in Odisha, India. Gut Pathog. 2017;9:16.
- [Google Scholar]
- Norwalk-like virus and bacterial pathogens associated with cases of gastroenteritis onboard a US Navy ship. Am J Trop Med Hyg. 1999;61:904-8.
- [Google Scholar]
- Simultaneous isolation of verotoxin-producing strains of Escherichia coli O128:H2 and viruses in gastroenteritis outbreaks. Comp Immunol Microbiol Infect Dis. 2001;24:135-42.
- [Google Scholar]
- Synergistic effects between rotavirus and coinfecting pathogens on diarrheal disease: Evidence from a community-based study in Northwestern Ecuador. Am J Epidemiol. 2012;176:387-95.
- [Google Scholar]
- Etiologic profile of acute diarrhea in children in São Paulo. J Pediatr (Rio J). 2002;78:31-8.
- [Google Scholar]
- Incidence and etiology of infantile diarrhea and major routes of transmission in Huascar, Peru. Am J Epidemiol. 1989;129:785-99.
- [Google Scholar]
- Occurrence of colonization factor antigens I & II in enterotoxigenic Escherichia coli associated diarrhoea in Iran & correlation with severity of disease. Indian J Med Res. 1992;95:115-20.
- [Google Scholar]
- Enteric pathogens in severe forms of acute gastroenteritis in Ghanaian children. Acta Paediatr Jpn Overseas Ed. 1996;38:672-6.
- [Google Scholar]
- Acute gastroenteritis surveillance through the National outbreak reporting system, United States. Emerg Infect Dis. 2013;19:1305-9.
- [Google Scholar]
- New real-time PCR detects prolonged norovirus excretion in highly immunosuppressed patients and children. J Clin Microbiol. 2009;47:2855-62.
- [Google Scholar]
- Quantitative detection of norovirus excretion in pediatric patients with cancer and prolonged gastroenteritis and shedding of norovirus. J Med Virol. 2008;80:1461-7.
- [Google Scholar]
- Is there a role for oral human immunoglobulin in the treatment for norovirus enteritis in immunocompromised patients? Pediatr Transplant. 2011;15:718-21.
- [Google Scholar]
- Noroviruses as a potential cause of protracted and lethal disease in immunocompromised patients. Clin Infect Dis. 2009;49:1069-71.
- [Google Scholar]
- Impact of norovirus/sapovirus-related diarrhea in renal transplant recipients hospitalized for diarrhea. Transplantation. 2011;92:61-9.
- [Google Scholar]
- Chronic norovirus infection in renal transplant recipients. Nephrol Dial Transplant. 2008;24:1051-3.
- [Google Scholar]
- Changing pattern of prevalence, genetic diversity, and mixed infections of viruses associated with acute gastroenteritis in pediatric patients in New Delhi, India. J Med Virol. 2018;90:469-76.
- [Google Scholar]
- Epidemiological, clinical, and molecular features of norovirus infections in western India. J Med Virol. 2009;81:922-32.
- [Google Scholar]
- Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J Med Virol. 2007;79:544-51.
- [Google Scholar]
- Giardiasis: A review on assemblage distribution and epidemiology in India. Indian J Gastroenterol. 2012;31:3-12.
- [Google Scholar]
- Gastrointestinal complications in renal transplant recipients. Am J Clin Pathol. 1986;86:161-7.
- [Google Scholar]
- Colorectal disease in liver allograft recipients – A clinicopathological study with follow-up. Eur J Gastroenterol Hepatol. 2002;14:231-6.
- [Google Scholar]







