Translate this page into:
Mass Spectrometric Identification of Urinary Biomarkers of Chronic Kidney Disease: A Proteomic-Related Preliminary Report
Corresponding author: Ravikumar Sambandam, Multi-Disciplinary Center for Biomedical Research, Aarupadai Veedu Medical College and Hospital, Vinayaka Missions Research Foundation (Deemed to be University), Kirumampakkam, Puducherry, India. E-mail: ravikumar.sambandam@avmc.edu.in
-
Received: ,
Accepted: ,
How to cite this article: Kademani SP, Nelaturi P, Kalidas SS, Ballambattu VB, Sambandam R. Mass Spectrometric Identification of Urinary Biomarkers of Chronic Kidney Disease: A Proteomic-Related Preliminary Report. Indian J Nephrol. 2025;35:253-8. doi: 10.25259/ijn_255_23
Abstract
Background
Chronic kidney disease (CKD) is a gradual loss of kidney function and has an increased prevalence rate worldwide. Our study was intended to identify potential biomarkers of progression using urine proteomics.
Materials and Methods
This preliminary study consisted of 32 patients with stage V CKD. Urine samples were subjected to liquid chromatography–mass spectrometry (LCMS), and the network of protein interaction was analyzed using STRING.
Results
A total of 135 proteins were identified, of which 35 were listed as candidates based on their clinical significance. Protein– protein interaction study provides novel insights into the functional constitution of the proteome, selecting urine as a source of biomarkers.
Conclusion
The present study observed that the potential markers such as EndoG, HPX, APN, AnxA1, Mic60, LONP1, and HYOU1 correlate with renal damage and its progression to CKD stage V.
Keywords
Biomarker
Chronic kidney disease
Protein-protein interaction
Urine proteome
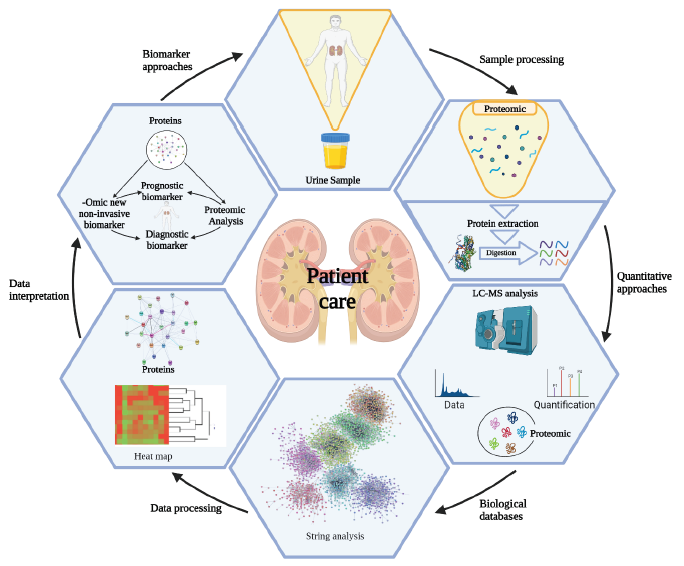
Introduction
Chronic kidney disease (CKD)1 has a global prevalence of 11–13%.2 Progression of CKD depends on the primary disease and also other influencing factors such as smoking, diet, or coexisting metabolic diseases.3 Inflammatory response associated with oxidative stress promotes renal damage by inducing necrosis, apoptosis, and fibrosis, which may play a role in the progression of CKD.4 Several markers of various risk factors of CKD can be measured in the tissue or serum to monitor the disease and its progression.
Investigation of alternative biofluids such as urine as a potential non-invasive biomarker5 would provide an easier means of disease monitoring. Urinary peptides identified through liquid chromatography–mass spectrometry (LCMS) could help in monitoring the disease progression and treatment response.
The study aims to identify a urinary proteome-based panel of novel protein biomarkers of CKD.
Materials and Methods
This preliminary study consisted of 32 patients with CKD stage V recruited at the Division of Nephrology, Department of General Medicine, Aarupadai Veedu Medical College and Hospital, Puducherry, India. The study was approved by the Institutional Human Ethics Committee of AVMC & H (IHEC No: AV/IEC/2021/010), and informed consent was obtained from each participant. The participants were asked to answer questionnaires regarding anthropometric measurements and recorded their laboratory and clinical examinations. The sample collection and preparation workflow are represented in Figure 1.
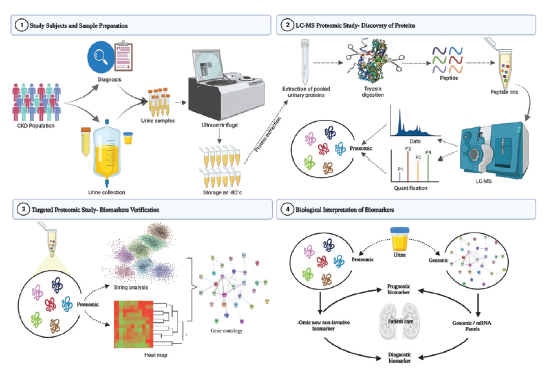
- Urine sample collection and preparation work flow.
Sample collection and preparation
Fresh urine samples were collected in the early morning, clean catch specimen into a sterile urine container (15 mL falcon tube), and immediately transported to the ice to prevent proteolysis and microbe contamination. Samples were processed within 30 min to remove cellular components by centrifugation at ≈3000 relative centrifugal forces (RCF) for 20 min at 4°C.6 The supernatant (5 mL each aliquot) was aliquoted and stored at -80°C.
Mass spectrometric analysis
The supernatants were concentrated and digested overnight with trypsin at 37°C. The precipitated protein pellets were re-dissolved in 120 µL of the initial mobile phase consisting of 0.1% formic acid in acetonitrile. The resulting peptides were analyzed using nano-LC-MS/MS-LTQ-orbitrap discovery (Thermo Scientific). Database searches were performed using the proteome discover software, UniProt, and Mascot search engines for the human database.
Protein–protein interaction analysis
The network of protein interaction was analyzed the using STRING software, based on the STRING database and gene ontology. Verified candidate proteins that were found in urine samples of CKD stage V were subjected to further protein–protein network analysis.
Results
The data on the baseline characteristics of the study subjects are presented in Table 1. We identified 135 proteins and segregated them based on gene ontology enrichment analysis, shown in Supplementary Data 1. Out of 135 proteins, 35 showed higher expression in urine [Table 2] and are involved in pathophysiological processes of kidneys such as protein sorting, ion and/or iron transport, metal binding, hemostasis, protease and antiprotease activity, transcriptional regulation, stress response, and apoptosis. We chose 10 candidates, namely, CD59, ANPEP, APOH, HPX, LMAN2, ANXA4, ANXA5, TPP1, ANXA11, and ATP1A1, having significance in the progression of CKD to end-stage renal disease.
| Parameters | Mean ± SD (CKD stage-V [n = 32]) |
|---|---|
| Age (years) | 55.77 ± 8.04 |
| Body mass index (BMI) (kg/m2) | 22.00 ± 4.0 |
| Blood urea (mg/dL) | 105.81 ± 23.75 |
| Serum creatinine (mg/dL) | 9.21 ± 2.78 |
| Uric acid (mg/dL) | 9.42 ± 1.47 |
| 24 h Creatinine clearance (mL/min) | 165.10 ± 40.11 |
| Creatinine clearance (Cockcroft–Gault equation) (mL/min) | 15.63 ± 1.83 |
| Proteinuria (g/24 h) | 1.48 ± 0.28 |
| Cystatin-C (mg/L) | 2.55 ± 0.40 |
| eGFR (mL/min/1.73 m2) | 8.93 ± 1.23 |
| Albumin mg/dL | 4.05 ± 0.64 |
| Urine: ACR (mg/g) | 271.59 ± 61.31 |
| Ammonia (µmol/L) | 53.16 ± 19.37 |
| pH | 7.31 ± 0.58 |
| Specific gravity | 1.020 ± 0.01 |
| Serum electrolytes | |
| Calcium (mg/dl) | 7.57 ± 1.48 |
| Phosphorous (mg/dl) | 6.69 ± 2.58 |
| Sodium (mmol/L) | 140.87 ± 4.92 |
| Potassium (mmol/L) | 4.73 ± 0.66 |
SD: Standard deviation, CKD: Chronic kidney disease, eGFR: Estimated glomerular filtration rate, ACR: Albumin to creatinine ratio.
| Protein (Uniprot ID) | Synonym | pI | Molecular weight (kDa) | Gene | Anatomical entity | Theoretical expression | Function | Sequence |
|---|---|---|---|---|---|---|---|---|
| CD59 glycoprotein (P13987) | MIC11, MIN1, MIN2, MIN3, MSK21 | 6.02 | 14.17 | CD59 | Renal medulla | 98.87 | Transport, sorting, protein transport | TAVNCSSDFDACLITK |
| Aminopeptidase N (P15144) | CD13 | 5.31 | 109.4 | ANPEP | Kidney | 98.07 | Protease activity, angiogenesis, differentiation, host-viral interaction | VVTVIAHELAHQWFGNLVTI |
| Annexin A1 (P04083) | ANX1, LPC1 | 6.57 | 38.71 | ANXA1 | Urethra | 98.19 | Inflammatory response, calcium/ phospholipid binding | QAWFIENEEQEYVQTVK |
| Progranulin (P28799) | GP88, PCDGF, PEP | 6.43 | 63.54 | GRN | Kidney | 97.31 | Integral membrane glycoprotein, immunity | DVECGEGHFCHDNQTCCR |
| Annexin A6 (P08133) | ANX6 | 5.41 | 75.87 | ANXA6 | Kidney | 93.8 | Regulate the release of Ca2+ | EDAQVAAEILEIADTPSGDK |
| Sodium/potassium-transporting ATPase subunit alpha-1 (P05023) | 5.33 | 112.8 | ATP1A1 | Kidney | 99.54 | Sodium/potassium transport | DAFQNAYLELGGLGER | |
| Peroxiredoxin-1 (Q06830) | PAGA, PAGB, TDPX2 | 8.27 | 22.11 | PRDX1 | Kidney | 99.29 | Antioxidant | YVVFFFYPLDFTFVCPTEII AFSDR |
| Tripeptidyl-peptidase 1 (O14773) | CLN2 | 6.01 | 61.24 | TPP1 | Renal glomerulus | 97.09 | Tripeptidyl-peptidase I activity, metal binding | FLSSSPHLPPSSYFNASGR |
| Beta-2-glycoprotein 1 (P02749) | B2G1 | 8.34 | 38.29 | APOH | Renal glomerulus | 86.47 | Inhibitor of serine proteases, acute phase, blood coagulation, hemostasis | TFYEPGEEITYSCKPGYVSR |
| Vesicular integral-membrane protein VIP36 (Q12907) | C5orf8 | 6.46 | 40.22 | LMAN2 | Kidney | 93.99 | Heparin binding | LFQLMVEHTPDEESIDWTK |
| Hemopexin (P02790) | 6.55 | 51.67 | HPX | Kidney | 60.29 | Heme binding, transport | DGWHSWPIAHQWPQGPSAVDAAFSWEEK | |
| Annexin A11 (P50995) | ANX11 | 7.53 | 54.38 | ANXA11 | Kidney | 97.17 | Cell division | GFGTDEQAIIDCLGSR |
| Annexin A5 (P08758) | ANX5, ENX2, PP4 | 4.93 | 35.93 | ANXA5 | Urethra | 99.19 | Anticoagulant protein, hemostasis | DPDAGIDEAQVEQDAQALFQAGELK |
| Annexin A4 (P09525) | ANX4 | 5.83 | 35.88 | ANXA4 | Urethra | 97.72 | Exocytosis. calcium/phospholipid binding | AASGFNAMEDAQTLR |
| Glutaminase kidney isoform (O94925) | GLS1, KIAA0838 | 7.85 | 73.46 | GLS | Kidney | 97.1 | Acid-base homeostasis | ILQEYQVQYTPQGDSDNGK |
| MICOS complex subunit MIC60 (Q16891) | HMP, MIC60, MINOS2 | 6.08 | 83.67 | IMMT | Renal medulla | 96.32 | Mitochondrial cristae morphology, host-viral interaction | LSQEQVDNFTLDINTAYAR |
| Lon protease homolog (P36776) | PRSS15 | 6.01 | 106.4 | LONP1 | Kidney epithelium | 85.64 | ATP-binding | EIFDIAFPDEQAEALAVER |
| Hypoxia-inducible factor 3-alpha (Q9Y2N7 | BHLHE17, MOP7, PASD7 | 5.67 | 72.43 | HIF3A | Kidney | 73.12 | Transcriptional regulator, angiogenesis, stress response, apoptosis, transcription regulation | DTEAVETDLDIAQDADALDL |
| Protein (Uniprot ID) | Synonym | pI | Molecular weight (kDa) | Gene | Anatomical entity | Theoretical expression | Function | Sequence |
| Peroxiredoxin-4 (Q13162) | 5.86 | 30.53 | PRDX4 | Renal glomerulus | 93.48 | Antioxidant | YLVFFFYPLDFTFVCPTEII AFGDR | |
| Hypoxia up-regulated protein 1 (Q9Y4L1) | GRP170, ORP150 | 5.16 | 111.3 | HYOU1 | Kidney | 94.29 | Cytoprotective, chaperone, stress response | LIPEMDQIFTEVEMTTLEK |
| Chloride intracellular channel protein 1 (O00299) | G6, NCC27 | 5.09 | 26.92 | CLIC1 | Kidney | 98.89 | Voltage-gated channel, chloride channel | FLDGNELTLADCNLLPK |
| Lysosome-associated membrane glycoprotein 1 (P11279) | CD107a | 9 | 44.88 | LAMP1 | Renal glomerulus | 99.92 | Lysosome biogenesis, autophagy, cholesterol homeostasis, Host-viral interaction | YSVQLMSFVYNLSDTHLFPN ASSK |
| L-xylulose reductase (Q7Z4W1) | SDR20C1 | 8.33 | 25.91 | DCXR | Kidney | 98.65 | Osmoregulation, carbohydrate metabolism, glucose metabolism, xylose metabolism | ALGSVGPVDLLVNNAAVALL QPFLEVTK |
| Radixin (P35241) | 6.03 | 68.56 | RDX | Urethra | 94.32 | Actin binding | VTTMDAELEFAIQPNTTGK | |
| Myosin-9 (P35579) | 5.5 | 226.5 | MYH9 | Kidney | 95 | Actin binding, calmodulin binding, cell adhesion | LQQLFNHTMFILEQEEYQR | |
| EH domain-containing protein 4 (Q9H223) | HCA10, HCA11, PAST4 | 6.32 | 61.17 | EHD4 | Urethra | 91.13 | ATP- and membrane binding protein | LDGYELPSSLPPHLVPPSHR |
| Neutrophil gelatinase-associated lipocalin (P80188) | HNL, NGAL | 9.02 | 22.58 | LCN2 | Kidney | 75.41 | Renal development, apoptosis, immunity, iron transport, ion transport | TFVPGCQPGEFTLGNIK |
| Annexin A3 (P12429) | ANX3 | 5.62 | 36.37 | ANXA3 | Kidney | 80.25 | Anticoagulant, phospholipase A2 inhibitor | GELSGHFEDLLLAIVNCVR |
| Serpin B6 (P35237) | PI6, PTI | 5.18 | 42.59 | SERPINB6 | Kidney | 96.52 | Mitochondrial membrane ATP synthase (F1F0 ATP synthase of complex V), hydrogen ion transport | TYIGEIFTQILVLPYVGK |
| Calnexin (P27824) | 4.46 | 67.56 | CANX | Cortex of kidney | 98.64 | Calcium binding protein, chaperone | WKPPMIDNPSYQGIWKPR | |
| Leukocyte elastase inhibitor (P30740) | ELANH2, MNEI, PI2 | 5.9 | 42.74 | SERPINB1 | Kidney | 87.3 | Transcriptional regulator | TYGADLASVDFQHASEDAR |
| Amyloid-beta precursor protein (P05067) | A4, AD1 | 4.73 | 86.94 | APP | Kidney epithelium | 98.63 | Heparin-binding, apoptosis, cell adhesion, endocytosis | LALENYITALQAVPPRPR |
| Transmembrane emp24 domain-containing protein 9 (Q9BVK6) | GP25L2 | 7.81 | 27.27 | TMED9 | Renal glomerulus | 98.34 | Protein recruitment, mRNA transport, translocation, transport | VHLDIQVGEHANDYAEIAAK |
| Endoplasmic reticulum chaperone BiP (P11021) | HSPA5 | 5.07 | 72.33 | GRP78 | Renal medulla | 99.08 | Chaperone, apoptosis, cell proliferation | GINPDEAVAYGAAVQAGVLS |
| Stress-70 protein, mitochondrial (P38646) | GRP75, HSPA9B, mt-HSP70 | 5.87 | 73.68 | HSPA9 | Renal glomerulus | 95.39 | Chaperone | STNGDTFLGGEDFDQALLR |
LC-MS: Liquid chromatography-mass spectrometry.
A protein interaction network was constructed to evaluate any functional and/or physical interactions between the urinary protein candidates for 135 proteins using STRING and functional ontology (GO) enrichment analysis [Figure 2a-2b, Figure S1 and S2]. This analysis revealed that the majority of urinary protein candidates were membrane proteins associated with biological processes of heme homeostasis, protein trafficking and transport, and many of them were involved in the process of gene expression and cell signaling. Gene ontology analysis provides a comprehensive resource related to genes and gene products. The figure represents the list of proteins; proteins and peptides are interconnected in PPI networks, which can be altered in diseased conditions [Figure 2].
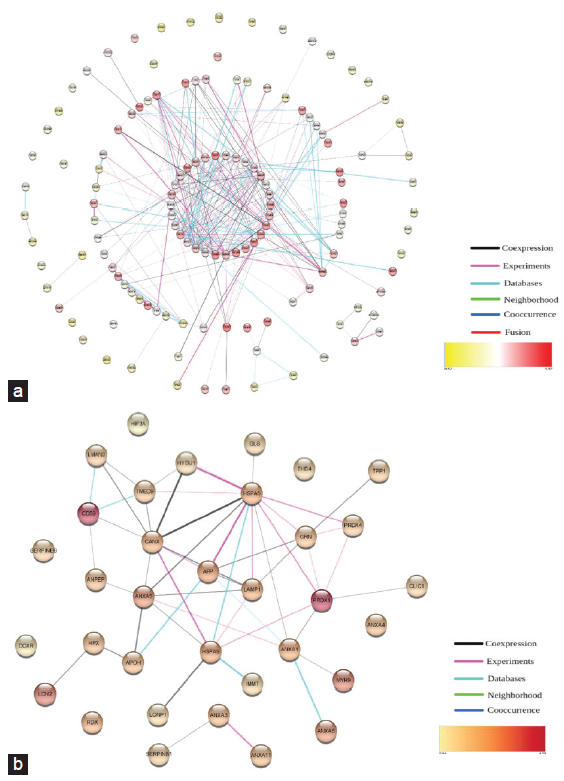
- Protein interaction network analysis. (a) Protein interaction analysis of 135 proteins identified in urine. (b) Protein interaction analysis of 35 proteins identified in urine.
Glycoproteins (CD59, HPX, LMAN2, and APOH) showed increased expression in urine. SERPINA1 identified in urine samples of patients with CKD acts on SERPINE1, which is a potent fibrosis-promoting glycoprotein. Endonucleases are unique and may be assembled with a variety of enzymes typically associated with DNA damage and/or repair (e.g. XRCC3, FANCG, ATP23, FEN1, APTX, and SFN) [Supplementary Data 1]. Results showed the presence of the most active apoptotic endonucleases of the kidneys such as DNAse 1 and EndoG in urine. We identified F- and P-type ATPases in urine, which include ATP1A1, ATP2A3, ATP2B2, ATP7B, ATP2A1, and ATP5IF1; these are involved in sodium/potassium, calcium, and copper transport. ATP5IF1 acts as an inhibitor of endogenous F1F(o)-ATPase. The zinc finger proteins are involved in transcriptional regulation. These zinc finger proteins (e.g. SNAI3, ZNF621, ZNF511, ZNF205, ZNF331, ZNF787, and DNLZ) have a promising role in the development of podocyte damage or glomerular diseases.
Discussion
Proteomics provide good tools with clinically relevant diagnosis and prognosis models as translational research. Our study showed 35 urinary proteins with increased expression in urine in patients with CKD stage V. These proteins are involved in various physiological and pathophysiological activities of the kidneys including cell signaling, inflammation, oxidative stress, hypoxia, DNA damage/repair mechanism, cell cycle arrest, mitochondrial dysfunction, heme homeostasis, and chaperons. CD59, a glycosylphosphatidylinositol (GPI) is composed of a phosphatidylinositol group linked through carbohydrate groups acts as an anchored glycoprotein and is ubiquitously expressed on epithelial, endothelial and mesangial cells of glomeruli. A study used anti-CD59 to neutralize its activity observed reduced renal function with severe endothelial damage with increased platelet and also fibrin deposition in glomeruli.7 Scavenging of free heme with hemopexin (HPX) is expected to mitigate heme-induced cell injury. Increased free heme concentration and HPX deficiency indicate that increased expression of CD59 could defend against complement-dependent renal injury.8 Aminopeptidase N (APN) is identical to CD13, belongs to type I metalloproteases, and has a role in metastasis, tumor invasion, and angiogenesis. The potential use of 99mTc-probestin SPECT as a new technique is imaging kidney APN expression.9 Increased urinary APN levels associated with microalbuminuria indicate tubular damage in diabetic nephropathy (DN).10
Annexins (Anx) can mediate a varied range of cellular processes including endo-, exocytosis, and membrane-cytoskeletal organization.11 AnxA1 in renal tissue is negatively correlated with the degree of tubular damage.12 AnxA1 levels correlated with normal kidney outcomes and showed higher expression during DN.13 AnxA4 regulates adenylyl cyclase type 5 and affects β-adrenoceptor/cAMP-dependent signaling.14 AnxA6 has a role in integrin recycling, aberrant Src, and FAK signaling, ECM cell surface delivery15 and IL-2 homeostasis-related T-cell proliferation.16 Progranulin (PGRN) is a cysteine-rich secretory protein used to predict the early signs of podocyte injury in DN17 and also indicates an endotoxin-induced septic acute kidney injury.18
Sodium/potassium-transporting ATPase (Na/K-ATPase) is an important ion pump of P-type ATPase. Reduced Na/K-ATPase α1 causes deficiency in signaling protein Src and NFkB activation. A study identified sodium/potassium-transporting ATPase subunit gamma in urine using capillary electrophoresis-coupled mass spectrometry in IgA nephropathy patients.19 Na/K-ATPase involved in CD40 receptor activation in the renal tubular epithelium during pathogenesis.20 Tripeptidyl peptidase-I (TPP-I) is a lysosomal serine proteinase expressed in the kidneys. Urine TPPI was significantly increased in patients with CKD.21 Peroxiredoxins (PRDXs) is an antioxidant enzyme key regulator of apoptosis and cell survival, it can intermediate during H2O2-induced activation of c-Abl/MST1/FOXO signaling.22 Mic060 is an important factor involved in the regulation of cell apoptosis, mitochondrial biogenesis, and oxidative stress.23 A study reported significantly decreased Mic60 levels in hemodialysis patients.24
Lon protease homolog, mitochondrial (LONP1) is a serine peptidase that protects the kidneys by inhibiting mitochondrial dysfunction and podocyte apoptosis.25 Radixin is one of the membrane-cytoskeletal crosslinkers, and increased levels of radixin were observed in dialysis patients.26 Limitations of this preliminary study are smaller sample size and that the proteins identified herein require further external validation.
Protein identification using LC-MS/MS allowed us to identify 135 proteins expressed in the urine of CKD patients. Possibly further studies on EndoG, HPX, APN, AnxA1, Mic60, LONP1, and HYOU1 potential markers will enable a better understanding of the mechanism of CKD and may provide novel opportunities for therapy. Sufficiently powered cohort studies are required to confirm the usefulness of these candidates to help improve understanding of CKD progression.
Acknowledgments
The authors are thankful to all participants involved in this study, and C-CAMP, Bengaluru, for carrying out the LC-MS/MS study.
Conflicts of interest
There are no conflicts of interest.
References
- Physiology, renal. In: StatPearls. Treasure Island (FL): StatPearls Publishing; 2019.
- [Google Scholar]
- The global burden of chronic kidney disease: Estimates, variability and pitfalls. Nat Rev Nephrol. 2017;13:104-14.
- [CrossRef] [PubMed] [Google Scholar]
- Diabetes trends in Latin America. Diabetes Metab Res Rev. 2002;18:S27-31.
- [CrossRef] [PubMed] [Google Scholar]
- Chronic kidney disease as oxidative stress- and inflammatory-mediated cardiovascular disease. Antioxidants (Basel). 2020;9:752.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850-62.
- [CrossRef] [PubMed] [Google Scholar]
- Urine sample preparation and protein profiling by two-dimensional electrophoresis and matrix-assisted laser desorption ionization time of flight mass spectroscopy. Methods Mol Biol. 2008;428:141-57.
- [CrossRef] [PubMed] [Google Scholar]
- CD59 protects glomerular endothelial cells from immune-mediated thrombotic microangiopathy in rats. J Am Soc Nephrol. 1998;9:590-7.
- [CrossRef] [PubMed] [Google Scholar]
- Hemopexin modulates expression of complement regulatory proteins in rat glomeruli. Current Issues Mol Biol. 2021;43:1081-9.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Evaluation of (99m) Tc-probestin SPECT as a novel technique for noninvasive imaging of kidney aminopeptidase N expression. Mol Pharm. 2014;11:2948-53.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Diagnostic value of the aminopeptidase N, N-acetyl-beta-D-glucosaminidase and dipeptidylpeptidase IV in evaluating tubular dysfunction in patients with glomerulopathies. Renal Fail. 2008;30:896-903.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of AnnexinA1 as an endogenous regulator of RhoA, and its role in the pathophysiology and experimental therapy of type-2 diabetes. Front Immunol. 2019;10:571.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Annexin A1 alleviates kidney injury by promoting the resolution of inflammation in diabetic nephropathy. Kidney Int. 2021;100:107-21.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Annexin A4 is a novel direct regulator of adenylyl cyclase type 5. FASEB J. 2015;29:3773-87.
- [CrossRef] [PubMed] [Google Scholar]
- Annexin A6-A multifunctional scaffold in cell motility. Cell Adh Migr. 2017;11:288-304.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Annexin A6 regulates interleukin-2-mediated T-cell proliferation. Immunol cell Biol. 2016;94:543-53.
- [CrossRef] [PubMed] [Google Scholar]
- Progranulin alleviates podocyte injury via regulating CAMKK/AMPK-mediated autophagy under diabetic conditions. J Mol Med (Berl). 2019;97:1507-20.
- [CrossRef] [PubMed] [Google Scholar]
- Progranulin protects against endotoxin-induced acute kidney injury by downregulating renal cell death and inflammatory responses in mice. Int Immunopharmacol. 2016;38:409-19.
- [CrossRef] [PubMed] [Google Scholar]
- Urine proteomics for prediction of disease progression in patients with IgA nephropathy. Nephrol Dial Transplantat. 2021;37:42-52.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Ion channels as a therapeutic target for renal fibrosis. Front Physiol. 2022;13:1019028.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Quantitative planar array screen of 1000 proteins uncovers novel urinary protein biomarkers of lupus nephritis. Ann Rheum Dis. 2020;79:1349-61.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Peroxiredoxin 1-an antioxidant enzyme in cancer. J Cell Mol Med. 2017;21:193-202.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Mitochondrial inner membrane protein, Mic60/mitofilin in mammalian organ protection. J Cell Physiol. 2019;234:3383-93.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The role of mitofilin in left ventricular hypertrophy in hemodialysis patients. Ren Fail. 2018;40:252-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Reduced Lon protease 1 expression in podocytes contributes to the pathogenesis of podocytopathy. Kidney Int. 2021;99:854-69.
- [CrossRef] [PubMed] [Google Scholar]
- A role for the membrane proteome in human chronic kidney disease erythrocytes. Transl Res. 2012;160:374-83.
- [CrossRef] [PubMed] [Google Scholar]







